Metin Nafi Gurcan
Assessing Concordance between RNA-Seq and NanoString Technologies in Ebola-Infected Nonhuman Primates Using Machine Learning
Oct 30, 2024Abstract:This study evaluates the concordance between RNA sequencing (RNA-Seq) and NanoString technologies for gene expression analysis in non-human primates (NHPs) infected with Ebola virus (EBOV). We performed a detailed comparison of both platforms, demonstrating a strong correlation between them, with Spearman coefficients for 56 out of 62 samples ranging from 0.78 to 0.88, with a mean of 0.83 and a median of 0.85. Bland-Altman analysis further confirmed high consistency, with most measurements falling within 95% confidence limits. A machine learning approach, using the Supervised Magnitude-Altitude Scoring (SMAS) method trained on NanoString data, identified OAS1 as a key marker for distinguishing RT-qPCR positive from negative samples. Remarkably, when applied to RNA-Seq data, OAS1 also achieved 100% accuracy in differentiating infected from uninfected samples using logistic regression, demonstrating its robustness across platforms. Further differential expression analysis identified 12 common genes including ISG15, OAS1, IFI44, IFI27, IFIT2, IFIT3, IFI44L, MX1, MX2, OAS2, RSAD2, and OASL which demonstrated the highest levels of statistical significance and biological relevance across both platforms. Gene Ontology (GO) analysis confirmed that these genes are directly involved in key immune and viral infection pathways, reinforcing their importance in EBOV infection. In addition, RNA-Seq uniquely identified genes such as CASP5, USP18, and DDX60, which play key roles in immune regulation and antiviral defense. This finding highlights the broader detection capabilities of RNA-Seq and underscores the complementary strengths of both platforms in providing a comprehensive and accurate assessment of gene expression changes during Ebola virus infection.
Machine Learning-Based Analysis of Ebola Virus' Impact on Gene Expression in Nonhuman Primates
Jan 22, 2024
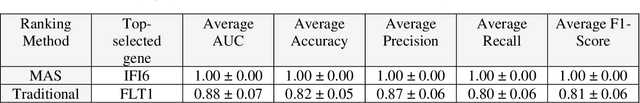

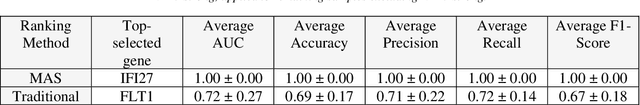
Abstract:This study introduces the Supervised Magnitude-Altitude Scoring (SMAS) methodology, a machine learning-based approach, for analyzing gene expression data obtained from nonhuman primates (NHPs) infected with Ebola virus (EBOV). We utilize a comprehensive dataset of NanoString gene expression profiles from Ebola-infected NHPs, deploying the SMAS system for nuanced host-pathogen interaction analysis. SMAS effectively combines gene selection based on statistical significance and expression changes, employing linear classifiers such as logistic regression to accurately differentiate between RT-qPCR positive and negative NHP samples. A key finding of our research is the identification of IFI6 and IFI27 as critical biomarkers, demonstrating exceptional predictive performance with 100% accuracy and Area Under the Curve (AUC) metrics in classifying various stages of Ebola infection. Alongside IFI6 and IFI27, genes, including MX1, OAS1, and ISG15, were significantly upregulated, highlighting their essential roles in the immune response to EBOV. Our results underscore the efficacy of the SMAS method in revealing complex genetic interactions and response mechanisms during EBOV infection. This research provides valuable insights into EBOV pathogenesis and aids in developing more precise diagnostic tools and therapeutic strategies to address EBOV infection in particular and viral infection in general.
Machine Learning Based Analytics for the Significance of Gait Analysis in Monitoring and Managing Lower Extremity Injuries
Sep 27, 2023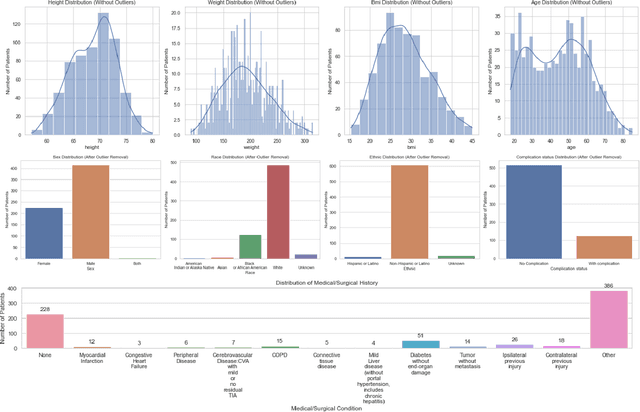
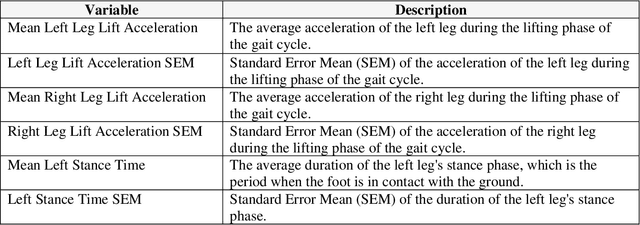
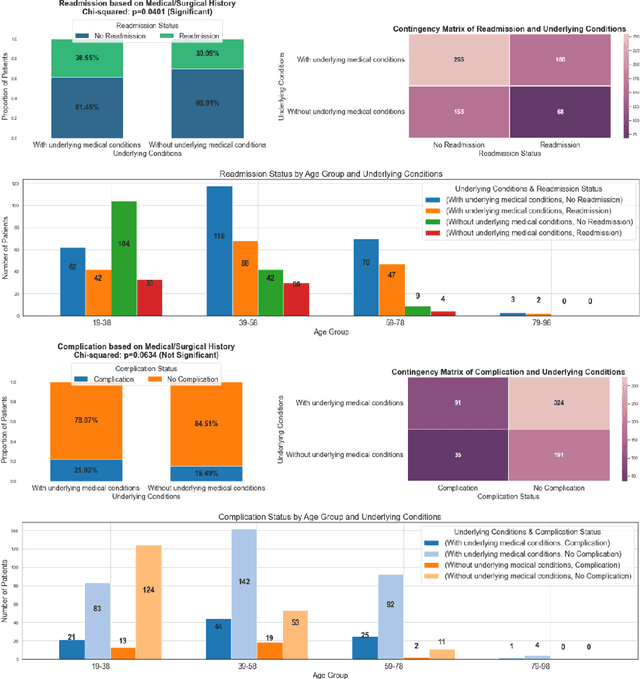

Abstract:This study explored the potential of gait analysis as a tool for assessing post-injury complications, e.g., infection, malunion, or hardware irritation, in patients with lower extremity fractures. The research focused on the proficiency of supervised machine learning models predicting complications using consecutive gait datasets. We identified patients with lower extremity fractures at an academic center. Patients underwent gait analysis with a chest-mounted IMU device. Using software, raw gait data was preprocessed, emphasizing 12 essential gait variables. Machine learning models including XGBoost, Logistic Regression, SVM, LightGBM, and Random Forest were trained, tested, and evaluated. Attention was given to class imbalance, addressed using SMOTE. We introduced a methodology to compute the Rate of Change (ROC) for gait variables, independent of the time difference between gait analyses. XGBoost was the optimal model both before and after applying SMOTE. Prior to SMOTE, the model achieved an average test AUC of 0.90 (95% CI: [0.79, 1.00]) and test accuracy of 86% (95% CI: [75%, 97%]). Feature importance analysis attributed importance to the duration between injury and gait analysis. Data patterns showed early physiological compensations, followed by stabilization phases, emphasizing prompt gait analysis. This study underscores the potential of machine learning, particularly XGBoost, in gait analysis for orthopedic care. Predicting post-injury complications, early gait assessment becomes vital, revealing intervention points. The findings support a shift in orthopedics towards a data-informed approach, enhancing patient outcomes.
Cross-attention-based saliency inference for predicting cancer metastasis on whole slide images
Sep 18, 2023



Abstract:Although multiple instance learning (MIL) methods are widely used for automatic tumor detection on whole slide images (WSI), they suffer from the extreme class imbalance within the small tumor WSIs. This occurs when the tumor comprises only a few isolated cells. For early detection, it is of utmost importance that MIL algorithms can identify small tumors, even when they are less than 1% of the size of the WSI. Existing studies have attempted to address this issue using attention-based architectures and instance selection-based methodologies, but have not yielded significant improvements. This paper proposes cross-attention-based salient instance inference MIL (CASiiMIL), which involves a novel saliency-informed attention mechanism, to identify breast cancer lymph node micro-metastasis on WSIs without the need for any annotations. Apart from this new attention mechanism, we introduce a negative representation learning algorithm to facilitate the learning of saliency-informed attention weights for improved sensitivity on tumor WSIs. The proposed model outperforms the state-of-the-art MIL methods on two popular tumor metastasis detection datasets, and demonstrates great cross-center generalizability. In addition, it exhibits excellent accuracy in classifying WSIs with small tumor lesions. Moreover, we show that the proposed model has excellent interpretability attributed to the saliency-informed attention weights. We strongly believe that the proposed method will pave the way for training algorithms for early tumor detection on large datasets where acquiring fine-grained annotations is practically impossible.
Attention2Minority: A salient instance inference-based multiple instance learning for classifying small lesions in whole slide images
Jan 18, 2023Abstract:Multiple instance learning (MIL) models have achieved remarkable success in analyzing whole slide images (WSIs) for disease classification problems. However, with regard to gigapixel WSI classification problems, current MIL models are often incapable of differentiating a WSI with extremely small tumor lesions. This minute tumor-to-normal area ratio in a MIL bag inhibits the attention mechanism from properly weighting the areas corresponding to minor tumor lesions. To overcome this challenge, we propose salient instance inference MIL (SiiMIL), a weakly-supervised MIL model for WSI classification. Our method initially learns representations of normal WSIs, and it then compares the normal WSIs representations with all the input patches to infer the salient instances of the input WSI. Finally, it employs attention-based MIL to perform the slide-level classification based on the selected patches of the WSI. Our experiments imply that SiiMIL can accurately identify tumor instances, which could only take up less than 1% of a WSI, so that the ratio of tumor to normal instances within a bag can increase by two to four times. It is worth mentioning that it performs equally well for large tumor lesions. As a result, SiiMIL achieves a significant improvement in performance over the state-of-the-art MIL methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge