Mehmet Efruz Karabulut
Sectioning of Biomedical Abstracts: A Sequence of Sequence Classification Task
Jan 18, 2022Abstract:Rapid growth of the biomedical literature has led to many advances in the biomedical text mining field. Among the vast amount of information, biomedical article abstracts are the easily accessible sources. However, the number of the structured abstracts, describing the rhetorical sections with one of Background, Objective, Method, Result and Conclusion categories is still not considerable. Exploration of valuable information in the biomedical abstracts can be expedited with the improvements in the sequential sentence classification task. Deep learning based models has great performance/potential in achieving significant results in this task. However, they can often be overly complex and overfit to specific data. In this project, we study a state-of-the-art deep learning model, which we called SSN-4 model here. We investigate different components of the SSN-4 model to study the trade-off between the performance and complexity. We explore how well this model generalizes to a new data set beyond Randomized Controlled Trials (RCT) dataset. We address the question that whether word embeddings can be adjusted to the task to improve the performance. Furthermore, we develop a second model that addresses the confusion pairs in the first model. Results show that SSN-4 model does not appear to generalize well beyond RCT dataset.
CU-UD: text-mining drug and chemical-protein interactions with ensembles of BERT-based models
Nov 11, 2021
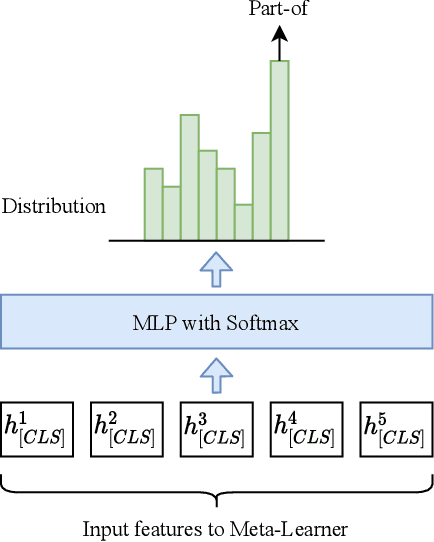
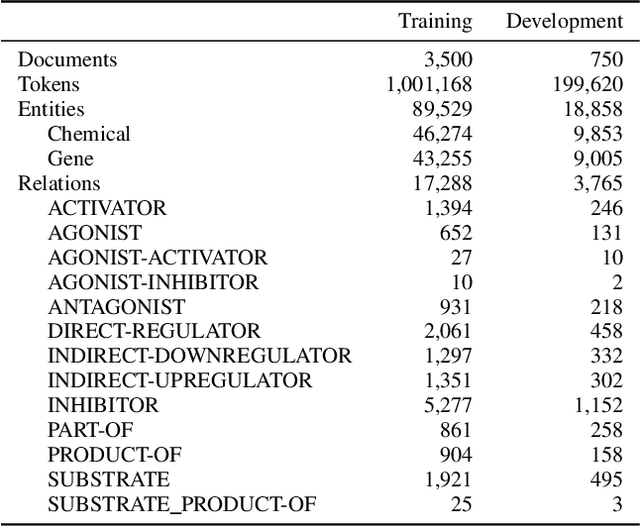
Abstract:Identifying the relations between chemicals and proteins is an important text mining task. BioCreative VII track 1 DrugProt task aims to promote the development and evaluation of systems that can automatically detect relations between chemical compounds/drugs and genes/proteins in PubMed abstracts. In this paper, we describe our submission, which is an ensemble system, including multiple BERT-based language models. We combine the outputs of individual models using majority voting and multilayer perceptron. Our system obtained 0.7708 in precision and 0.7770 in recall, for an F1 score of 0.7739, demonstrating the effectiveness of using ensembles of BERT-based language models for automatically detecting relations between chemicals and proteins. Our code is available at https://github.com/bionlplab/drugprot_bcvii.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge