Md Manowarul Islam
A Bioinformatic Approach Validated Utilizing Machine Learning Algorithms to Identify Relevant Biomarkers and Crucial Pathways in Gallbladder Cancer
Oct 18, 2024
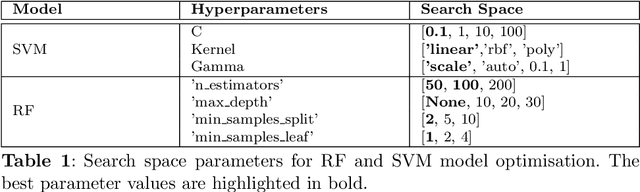
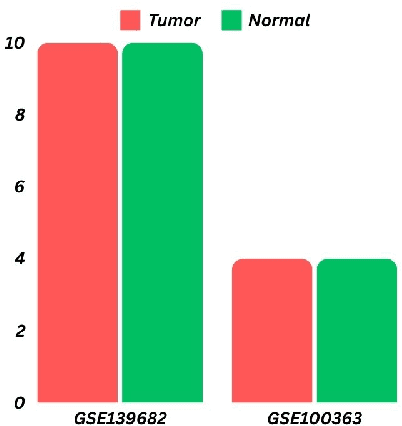

Abstract:Gallbladder cancer (GBC) is the most frequent cause of disease among biliary tract neoplasms. Identifying the molecular mechanisms and biomarkers linked to GBC progression has been a significant challenge in scientific research. Few recent studies have explored the roles of biomarkers in GBC. Our study aimed to identify biomarkers in GBC using machine learning (ML) and bioinformatics techniques. We compared GBC tumor samples with normal samples to identify differentially expressed genes (DEGs) from two microarray datasets (GSE100363, GSE139682) obtained from the NCBI GEO database. A total of 146 DEGs were found, with 39 up-regulated and 107 down-regulated genes. Functional enrichment analysis of these DEGs was performed using Gene Ontology (GO) terms and REACTOME pathways through DAVID. The protein-protein interaction network was constructed using the STRING database. To identify hub genes, we applied three ranking algorithms: Degree, MNC, and Closeness Centrality. The intersection of hub genes from these algorithms yielded 11 hub genes. Simultaneously, two feature selection methods (Pearson correlation and recursive feature elimination) were used to identify significant gene subsets. We then developed ML models using SVM and RF on the GSE100363 dataset, with validation on GSE139682, to determine the gene subset that best distinguishes GBC samples. The hub genes outperformed the other gene subsets. Finally, NTRK2, COL14A1, SCN4B, ATP1A2, SLC17A7, SLIT3, COL7A1, CLDN4, CLEC3B, ADCYAP1R1, and MFAP4 were identified as crucial genes, with SLIT3, COL7A1, and CLDN4 being strongly linked to GBC development and prediction.
A Hybrid Feature Fusion Deep Learning Framework for Leukemia Cancer Detection in Microscopic Blood Sample Using Gated Recurrent Unit and Uncertainty Quantification
Oct 18, 2024


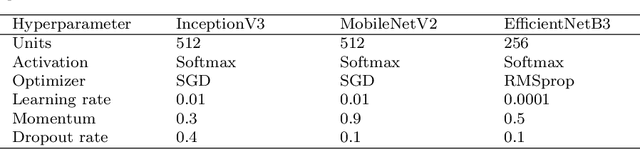
Abstract:Acute lymphoblastic leukemia (ALL) is the most malignant form of leukemia and the most common cancer in adults and children. Traditionally, leukemia is diagnosed by analyzing blood and bone marrow smears under a microscope, with additional cytochemical tests for confirmation. However, these methods are expensive, time consuming, and highly dependent on expert knowledge. In recent years, deep learning, particularly Convolutional Neural Networks (CNNs), has provided advanced methods for classifying microscopic smear images, aiding in the detection of leukemic cells. These approaches are quick, cost effective, and not subject to human bias. However, most methods lack the ability to quantify uncertainty, which could lead to critical misdiagnoses. In this research, hybrid deep learning models (InceptionV3-GRU, EfficientNetB3-GRU, MobileNetV2-GRU) were implemented to classify ALL. Bayesian optimization was used to fine tune the model's hyperparameters and improve its performance. Additionally, Deep Ensemble uncertainty quantification was applied to address uncertainty during leukemia image classification. The proposed models were trained on the publicly available datasets ALL-IDB1 and ALL-IDB2. Their results were then aggregated at the score level using the sum rule. The parallel architecture used in these models offers a high level of confidence in differentiating between ALL and non-ALL cases. The proposed method achieved a remarkable detection accuracy rate of 100% on the ALL-IDB1 dataset, 98.07% on the ALL-IDB2 dataset, and 98.64% on the combined dataset, demonstrating its potential for accurate and reliable leukemia diagnosis.
Hybridized Convolutional Neural Networks and Long Short-Term Memory for Improved Alzheimer's Disease Diagnosis from MRI Scans
Mar 08, 2024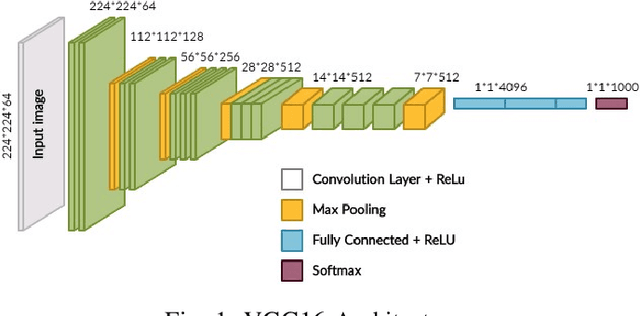
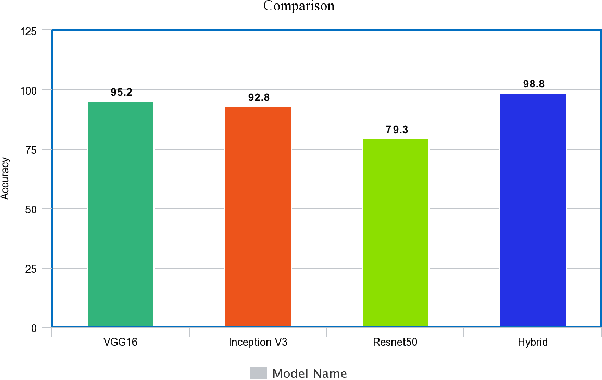
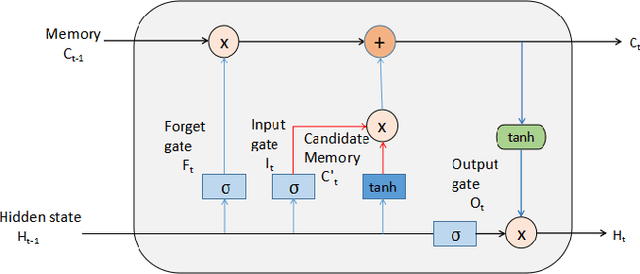
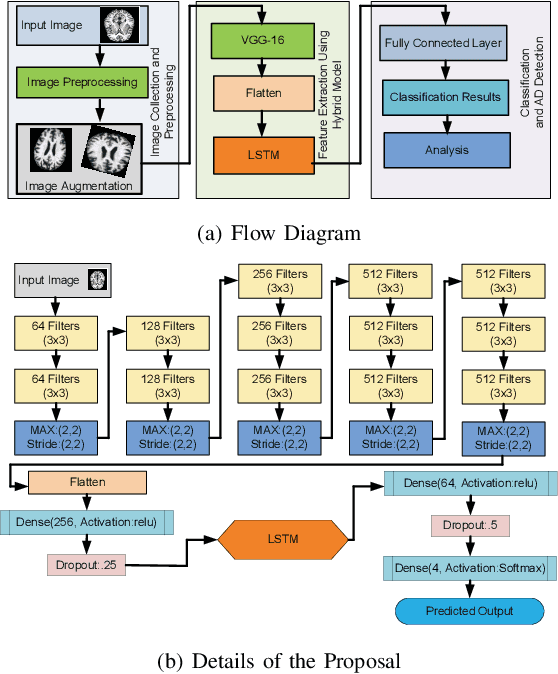
Abstract:Brain-related diseases are more sensitive than other diseases due to several factors, including the complexity of surgical procedures, high costs, and other challenges. Alzheimer's disease is a common brain disorder that causes memory loss and the shrinking of brain cells. Early detection is critical for providing proper treatment to patients. However, identifying Alzheimer's at an early stage using manual scanning of CT or MRI scans is challenging. Therefore, researchers have delved into the exploration of computer-aided systems, employing Machine Learning and Deep Learning methodologies, which entail the training of datasets to detect Alzheimer's disease. This study aims to present a hybrid model that combines a CNN model's feature extraction capabilities with an LSTM model's detection capabilities. This study has applied the transfer learning called VGG16 in the hybrid model to extract features from MRI images. The LSTM detects features between the convolution layer and the fully connected layer. The output layer of the fully connected layer uses the softmax function. The training of the hybrid model involved utilizing the ADNI dataset. The trial findings revealed that the model achieved a level of accuracy of 98.8%, a sensitivity rate of 100%, and a specificity rate of 76%. The proposed hybrid model outperforms its contemporary CNN counterparts, showcasing a superior performance.
Exploring Gene Regulatory Interaction Networks and predicting therapeutic molecules for Hypopharyngeal Cancer and EGFR-mutated lung adenocarcinoma
Feb 27, 2024Abstract:With the advent of Information technology, the Bioinformatics research field is becoming increasingly attractive to researchers and academicians. The recent development of various Bioinformatics toolkits has facilitated the rapid processing and analysis of vast quantities of biological data for human perception. Most studies focus on locating two connected diseases and making some observations to construct diverse gene regulatory interaction networks, a forerunner to general drug design for curing illness. For instance, Hypopharyngeal cancer is a disease that is associated with EGFR-mutated lung adenocarcinoma. In this study, we select EGFR-mutated lung adenocarcinoma and Hypopharyngeal cancer by finding the Lung metastases in hypopharyngeal cancer. To conduct this study, we collect Mircorarray datasets from GEO (Gene Expression Omnibus), an online database controlled by NCBI. Differentially expressed genes, common genes, and hub genes between the selected two diseases are detected for the succeeding move. Our research findings have suggested common therapeutic molecules for the selected diseases based on 10 hub genes with the highest interactions according to the degree topology method and the maximum clique centrality (MCC). Our suggested therapeutic molecules will be fruitful for patients with those two diseases simultaneously.
MLSTL-WSN: Machine Learning-based Intrusion Detection using SMOTETomek in WSNs
Feb 22, 2024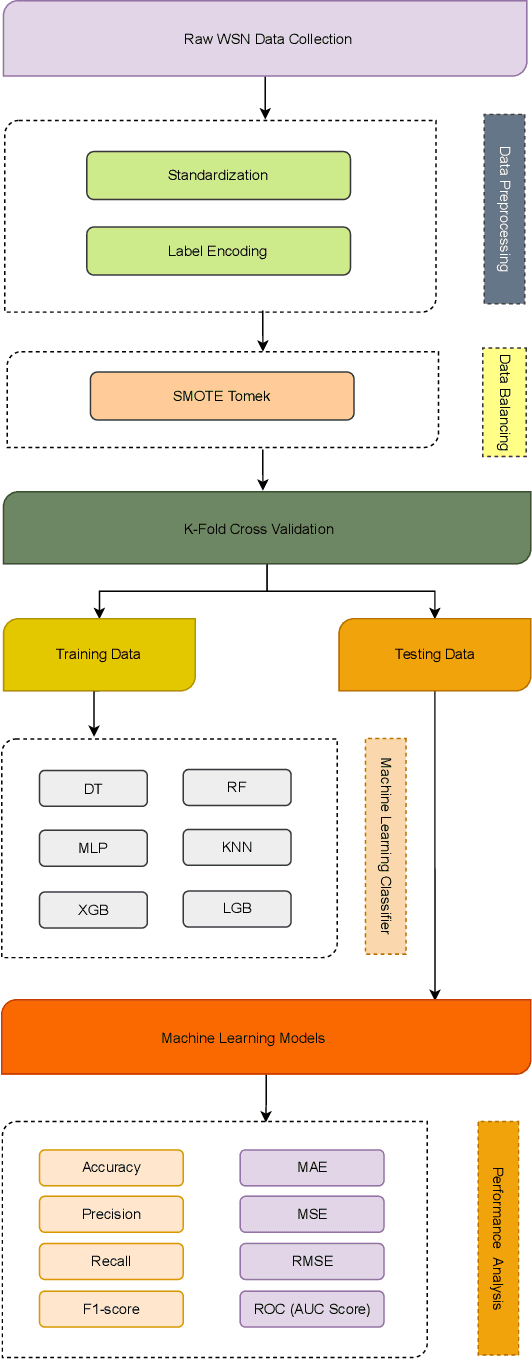

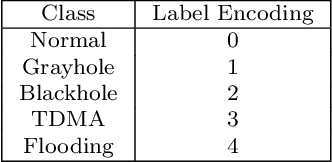
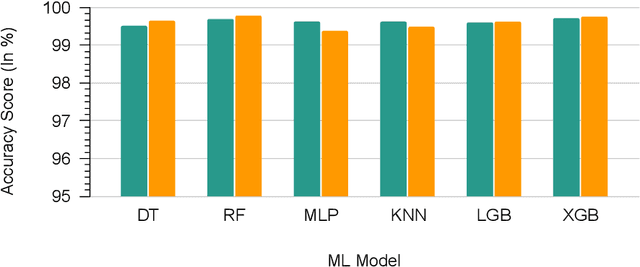
Abstract:Wireless Sensor Networks (WSNs) play a pivotal role as infrastructures, encompassing both stationary and mobile sensors. These sensors self-organize and establish multi-hop connections for communication, collectively sensing, gathering, processing, and transmitting data about their surroundings. Despite their significance, WSNs face rapid and detrimental attacks that can disrupt functionality. Existing intrusion detection methods for WSNs encounter challenges such as low detection rates, computational overhead, and false alarms. These issues stem from sensor node resource constraints, data redundancy, and high correlation within the network. To address these challenges, we propose an innovative intrusion detection approach that integrates Machine Learning (ML) techniques with the Synthetic Minority Oversampling Technique Tomek Link (SMOTE-TomekLink) algorithm. This blend synthesizes minority instances and eliminates Tomek links, resulting in a balanced dataset that significantly enhances detection accuracy in WSNs. Additionally, we incorporate feature scaling through standardization to render input features consistent and scalable, facilitating more precise training and detection. To counteract imbalanced WSN datasets, we employ the SMOTE-Tomek resampling technique, mitigating overfitting and underfitting issues. Our comprehensive evaluation, using the WSN Dataset (WSN-DS) containing 374,661 records, identifies the optimal model for intrusion detection in WSNs. The standout outcome of our research is the remarkable performance of our model. In binary, it achieves an accuracy rate of 99.78% and in multiclass, it attains an exceptional accuracy rate of 99.92%. These findings underscore the efficiency and superiority of our proposal in the context of WSN intrusion detection, showcasing its effectiveness in detecting and mitigating intrusions in WSNs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge