Maksuda Akter
An Integrated Deep Learning Model for Skin Cancer Detection Using Hybrid Feature Fusion Technique
Oct 18, 2024


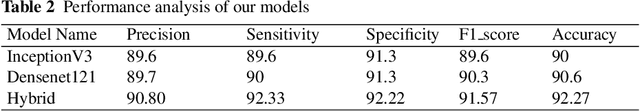
Abstract:Skin cancer is a serious and potentially fatal disease caused by DNA damage. Early detection significantly increases survival rates, making accurate diagnosis crucial. In this groundbreaking study, we present a hybrid framework based on Deep Learning (DL) that achieves precise classification of benign and malignant skin lesions. Our approach begins with dataset preprocessing to enhance classification accuracy, followed by training two separate pre-trained DL models, InceptionV3 and DenseNet121. By fusing the results of each model using the weighted sum rule, our system achieves exceptional accuracy rates. Specifically, we achieve a 92.27% detection accuracy rate, 92.33% sensitivity, 92.22% specificity, 90.81% precision, and 91.57% F1-score, outperforming existing models and demonstrating the robustness and trustworthiness of our hybrid approach. Our study represents a significant advance in skin cancer diagnosis and provides a promising foundation for further research in the field. With the potential to save countless lives through earlier detection, our hybrid deep-learning approach is a game-changer in the fight against skin cancer.
A Bioinformatic Approach Validated Utilizing Machine Learning Algorithms to Identify Relevant Biomarkers and Crucial Pathways in Gallbladder Cancer
Oct 18, 2024
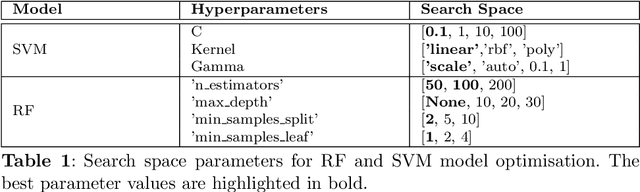
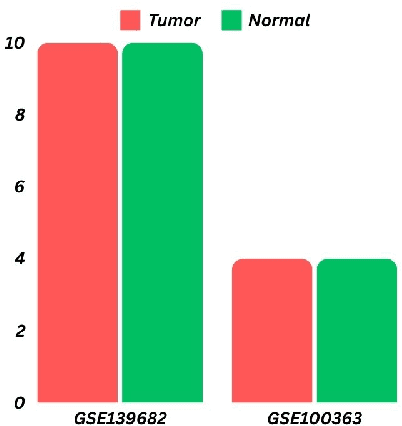

Abstract:Gallbladder cancer (GBC) is the most frequent cause of disease among biliary tract neoplasms. Identifying the molecular mechanisms and biomarkers linked to GBC progression has been a significant challenge in scientific research. Few recent studies have explored the roles of biomarkers in GBC. Our study aimed to identify biomarkers in GBC using machine learning (ML) and bioinformatics techniques. We compared GBC tumor samples with normal samples to identify differentially expressed genes (DEGs) from two microarray datasets (GSE100363, GSE139682) obtained from the NCBI GEO database. A total of 146 DEGs were found, with 39 up-regulated and 107 down-regulated genes. Functional enrichment analysis of these DEGs was performed using Gene Ontology (GO) terms and REACTOME pathways through DAVID. The protein-protein interaction network was constructed using the STRING database. To identify hub genes, we applied three ranking algorithms: Degree, MNC, and Closeness Centrality. The intersection of hub genes from these algorithms yielded 11 hub genes. Simultaneously, two feature selection methods (Pearson correlation and recursive feature elimination) were used to identify significant gene subsets. We then developed ML models using SVM and RF on the GSE100363 dataset, with validation on GSE139682, to determine the gene subset that best distinguishes GBC samples. The hub genes outperformed the other gene subsets. Finally, NTRK2, COL14A1, SCN4B, ATP1A2, SLC17A7, SLIT3, COL7A1, CLDN4, CLEC3B, ADCYAP1R1, and MFAP4 were identified as crucial genes, with SLIT3, COL7A1, and CLDN4 being strongly linked to GBC development and prediction.
A Hybrid Feature Fusion Deep Learning Framework for Leukemia Cancer Detection in Microscopic Blood Sample Using Gated Recurrent Unit and Uncertainty Quantification
Oct 18, 2024


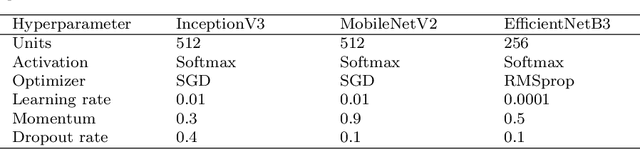
Abstract:Acute lymphoblastic leukemia (ALL) is the most malignant form of leukemia and the most common cancer in adults and children. Traditionally, leukemia is diagnosed by analyzing blood and bone marrow smears under a microscope, with additional cytochemical tests for confirmation. However, these methods are expensive, time consuming, and highly dependent on expert knowledge. In recent years, deep learning, particularly Convolutional Neural Networks (CNNs), has provided advanced methods for classifying microscopic smear images, aiding in the detection of leukemic cells. These approaches are quick, cost effective, and not subject to human bias. However, most methods lack the ability to quantify uncertainty, which could lead to critical misdiagnoses. In this research, hybrid deep learning models (InceptionV3-GRU, EfficientNetB3-GRU, MobileNetV2-GRU) were implemented to classify ALL. Bayesian optimization was used to fine tune the model's hyperparameters and improve its performance. Additionally, Deep Ensemble uncertainty quantification was applied to address uncertainty during leukemia image classification. The proposed models were trained on the publicly available datasets ALL-IDB1 and ALL-IDB2. Their results were then aggregated at the score level using the sum rule. The parallel architecture used in these models offers a high level of confidence in differentiating between ALL and non-ALL cases. The proposed method achieved a remarkable detection accuracy rate of 100% on the ALL-IDB1 dataset, 98.07% on the ALL-IDB2 dataset, and 98.64% on the combined dataset, demonstrating its potential for accurate and reliable leukemia diagnosis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge