A Bioinformatic Approach Validated Utilizing Machine Learning Algorithms to Identify Relevant Biomarkers and Crucial Pathways in Gallbladder Cancer
Paper and Code
Oct 18, 2024
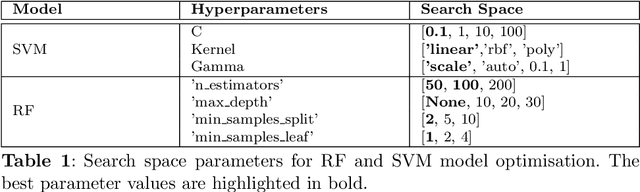
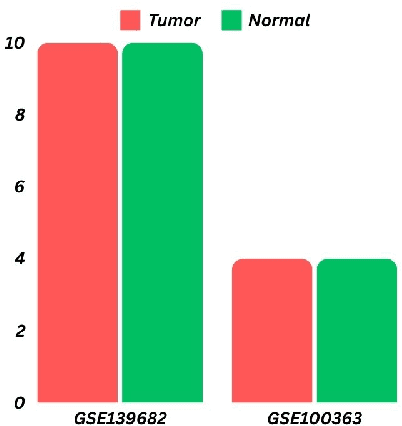

Gallbladder cancer (GBC) is the most frequent cause of disease among biliary tract neoplasms. Identifying the molecular mechanisms and biomarkers linked to GBC progression has been a significant challenge in scientific research. Few recent studies have explored the roles of biomarkers in GBC. Our study aimed to identify biomarkers in GBC using machine learning (ML) and bioinformatics techniques. We compared GBC tumor samples with normal samples to identify differentially expressed genes (DEGs) from two microarray datasets (GSE100363, GSE139682) obtained from the NCBI GEO database. A total of 146 DEGs were found, with 39 up-regulated and 107 down-regulated genes. Functional enrichment analysis of these DEGs was performed using Gene Ontology (GO) terms and REACTOME pathways through DAVID. The protein-protein interaction network was constructed using the STRING database. To identify hub genes, we applied three ranking algorithms: Degree, MNC, and Closeness Centrality. The intersection of hub genes from these algorithms yielded 11 hub genes. Simultaneously, two feature selection methods (Pearson correlation and recursive feature elimination) were used to identify significant gene subsets. We then developed ML models using SVM and RF on the GSE100363 dataset, with validation on GSE139682, to determine the gene subset that best distinguishes GBC samples. The hub genes outperformed the other gene subsets. Finally, NTRK2, COL14A1, SCN4B, ATP1A2, SLC17A7, SLIT3, COL7A1, CLDN4, CLEC3B, ADCYAP1R1, and MFAP4 were identified as crucial genes, with SLIT3, COL7A1, and CLDN4 being strongly linked to GBC development and prediction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge