Matthew Dowling
Meta-Dynamical State Space Models for Integrative Neural Data Analysis
Oct 07, 2024



Abstract:Learning shared structure across environments facilitates rapid learning and adaptive behavior in neural systems. This has been widely demonstrated and applied in machine learning to train models that are capable of generalizing to novel settings. However, there has been limited work exploiting the shared structure in neural activity during similar tasks for learning latent dynamics from neural recordings. Existing approaches are designed to infer dynamics from a single dataset and cannot be readily adapted to account for statistical heterogeneities across recordings. In this work, we hypothesize that similar tasks admit a corresponding family of related solutions and propose a novel approach for meta-learning this solution space from task-related neural activity of trained animals. Specifically, we capture the variabilities across recordings on a low-dimensional manifold which concisely parametrizes this family of dynamics, thereby facilitating rapid learning of latent dynamics given new recordings. We demonstrate the efficacy of our approach on few-shot reconstruction and forecasting of synthetic dynamical systems, and neural recordings from the motor cortex during different arm reaching tasks.
Large-scale variational Gaussian state-space models
Mar 03, 2024


Abstract:We introduce an amortized variational inference algorithm and structured variational approximation for state-space models with nonlinear dynamics driven by Gaussian noise. Importantly, the proposed framework allows for efficient evaluation of the ELBO and low-variance stochastic gradient estimates without resorting to diagonal Gaussian approximations by exploiting (i) the low-rank structure of Monte-Carlo approximations to marginalize the latent state through the dynamics (ii) an inference network that approximates the update step with low-rank precision matrix updates (iii) encoding current and future observations into pseudo observations -- transforming the approximate smoothing problem into an (easier) approximate filtering problem. Overall, the necessary statistics and ELBO can be computed in $O(TL(Sr + S^2 + r^2))$ time where $T$ is the series length, $L$ is the state-space dimensionality, $S$ are the number of samples used to approximate the predict step statistics, and $r$ is the rank of the approximate precision matrix update in the update step (which can be made of much lower dimension than $L$).
Linear Time GPs for Inferring Latent Trajectories from Neural Spike Trains
Jun 01, 2023



Abstract:Latent Gaussian process (GP) models are widely used in neuroscience to uncover hidden state evolutions from sequential observations, mainly in neural activity recordings. While latent GP models provide a principled and powerful solution in theory, the intractable posterior in non-conjugate settings necessitates approximate inference schemes, which may lack scalability. In this work, we propose cvHM, a general inference framework for latent GP models leveraging Hida-Mat\'ern kernels and conjugate computation variational inference (CVI). With cvHM, we are able to perform variational inference of latent neural trajectories with linear time complexity for arbitrary likelihoods. The reparameterization of stationary kernels using Hida-Mat\'ern GPs helps us connect the latent variable models that encode prior assumptions through dynamical systems to those that encode trajectory assumptions through GPs. In contrast to previous work, we use bidirectional information filtering, leading to a more concise implementation. Furthermore, we employ the Whittle approximate likelihood to achieve highly efficient hyperparameter learning.
Real-Time Variational Method for Learning Neural Trajectory and its Dynamics
May 18, 2023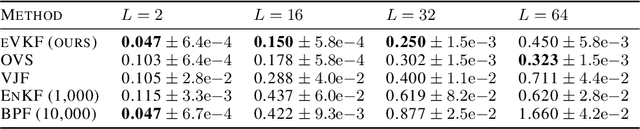

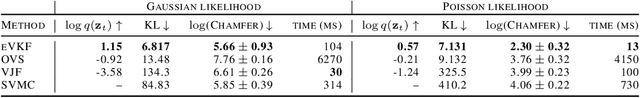

Abstract:Latent variable models have become instrumental in computational neuroscience for reasoning about neural computation. This has fostered the development of powerful offline algorithms for extracting latent neural trajectories from neural recordings. However, despite the potential of real time alternatives to give immediate feedback to experimentalists, and enhance experimental design, they have received markedly less attention. In this work, we introduce the exponential family variational Kalman filter (eVKF), an online recursive Bayesian method aimed at inferring latent trajectories while simultaneously learning the dynamical system generating them. eVKF works for arbitrary likelihoods and utilizes the constant base measure exponential family to model the latent state stochasticity. We derive a closed-form variational analogue to the predict step of the Kalman filter which leads to a provably tighter bound on the ELBO compared to another online variational method. We validate our method on synthetic and real-world data, and, notably, show that it achieves competitive performance
Hida-Matérn Kernel
Jul 15, 2021



Abstract:We present the class of Hida-Mat\'ern kernels, which is the canonical family of covariance functions over the entire space of stationary Gauss-Markov Processes. It extends upon Mat\'ern kernels, by allowing for flexible construction of priors over processes with oscillatory components. Any stationary kernel, including the widely used squared-exponential and spectral mixture kernels, are either directly within this class or are appropriate asymptotic limits, demonstrating the generality of this class. Taking advantage of its Markovian nature we show how to represent such processes as state space models using only the kernel and its derivatives. In turn this allows us to perform Gaussian Process inference more efficiently and side step the usual computational burdens. We also show how exploiting special properties of the state space representation enables improved numerical stability in addition to further reductions of computational complexity.
Non-parametric generalized linear model
Sep 02, 2020Abstract:A fundamental problem in statistical neuroscience is to model how neurons encode information by analyzing electrophysiological recordings. A popular and widely-used approach is to fit the spike trains with an autoregressive point process model. These models are characterized by a set of convolutional temporal filters, whose subsequent analysis can help reveal how neurons encode stimuli, interact with each other, and process information. In practice a sufficiently rich but small ensemble of temporal basis functions needs to be chosen to parameterize the filters. However, obtaining a satisfactory fit often requires burdensome model selection and fine tuning the form of the basis functions and their temporal span. In this paper we propose a nonparametric approach for jointly inferring the filters and hyperparameters using the Gaussian process framework. Our method is computationally efficient taking advantage of the sparse variational approximation while being flexible and rich enough to characterize arbitrary filters in continuous time lag. Moreover, our method automatically learns the temporal span of the filter. For the particular application in neuroscience, we designed priors for stimulus and history filters useful for the spike trains. We compare and validate our method on simulated and real neural spike train data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge