Matheus Palhares Viana
Bright 4B: Scaling Hyperspherical Learning for Segmentation in 3D Brightfield Microscopy
Dec 27, 2025Abstract:Label-free 3D brightfield microscopy offers a fast and noninvasive way to visualize cellular morphology, yet robust volumetric segmentation still typically depends on fluorescence or heavy post-processing. We address this gap by introducing Bright-4B, a 4 billion parameter foundation model that learns on the unit hypersphere to segment subcellular structures directly from 3D brightfield volumes. Bright-4B combines a hardware-aligned Native Sparse Attention mechanism (capturing local, coarse, and selected global context), depth-width residual HyperConnections that stabilize representation flow, and a soft Mixture-of-Experts for adaptive capacity. A plug-and-play anisotropic patch embed further respects confocal point-spread and axial thinning, enabling geometry-faithful 3D tokenization. The resulting model produces morphology-accurate segmentations of nuclei, mitochondria, and other organelles from brightfield stacks alone--without fluorescence, auxiliary channels, or handcrafted post-processing. Across multiple confocal datasets, Bright-4B preserves fine structural detail across depth and cell types, outperforming contemporary CNN and Transformer baselines. All code, pretrained weights, and models for downstream finetuning will be released to advance large-scale, label-free 3D cell mapping.
An argument in favor of strong scaling for deep neural networks with small datasets
Sep 25, 2018
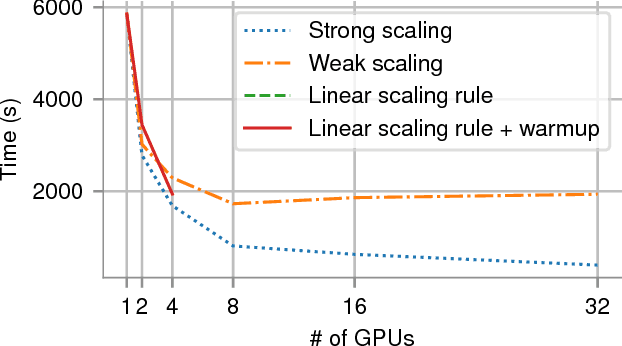
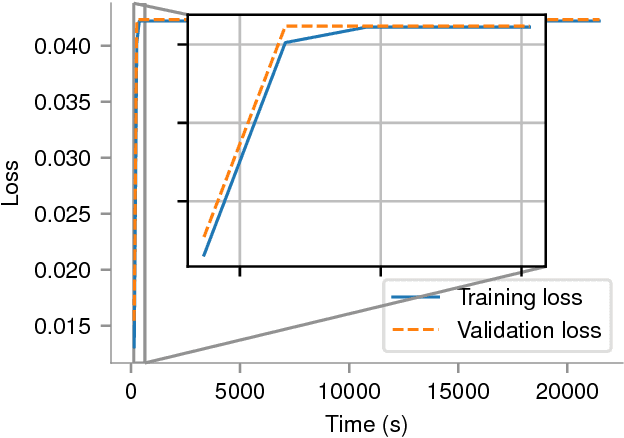
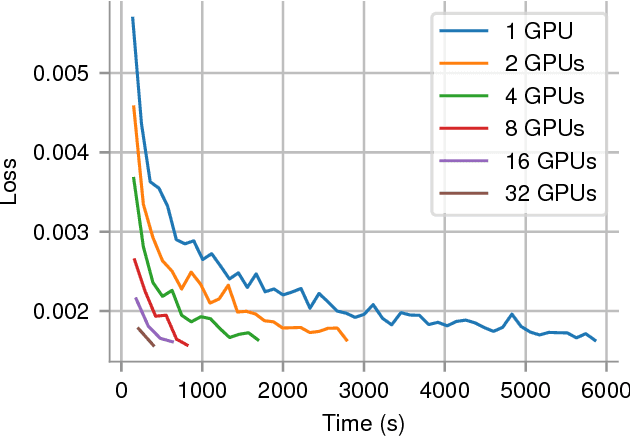
Abstract:In recent years, with the popularization of deep learning frameworks and large datasets, researchers have started parallelizing their models in order to train faster. This is crucially important, because they typically explore many hyperparameters in order to find the best ones for their applications. This process is time consuming and, consequently, speeding up training improves productivity. One approach to parallelize deep learning models followed by many researchers is based on weak scaling. The minibatches increase in size as new GPUs are added to the system. In addition, new learning rates schedules have been proposed to fix optimization issues that occur with large minibatch sizes. In this paper, however, we show that the recommendations provided by recent work do not apply to models that lack large datasets. In fact, we argument in favor of using strong scaling for achieving reliable performance in such cases. We evaluated our approach with up to 32 GPUs and show that weak scaling not only does not have the same accuracy as the sequential model, it also fails to converge most of time. Meanwhile, strong scaling has good scalability while having exactly the same accuracy of a sequential implementation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge