Maseud Rahgozar
DeepLink: A Novel Link Prediction Framework based on Deep Learning
Jul 27, 2018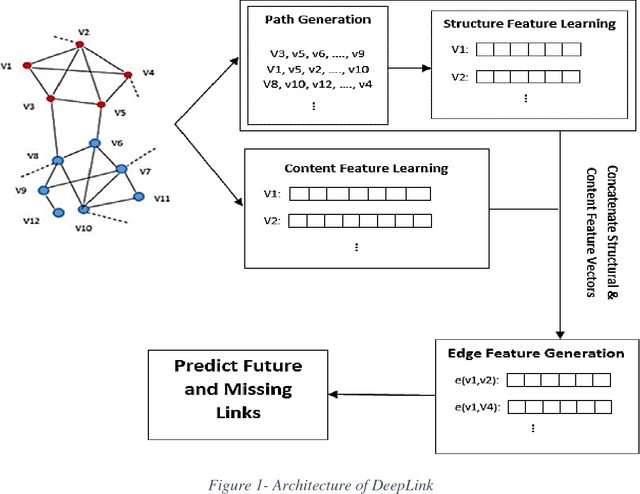
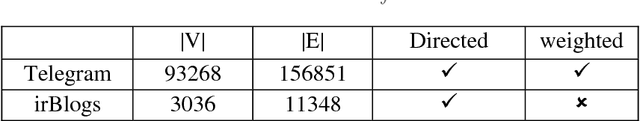
Abstract:Recently, link prediction has attracted more attentions from various disciplines such as computer science, bioinformatics and economics. In this problem, unknown links between nodes are discovered based on numerous information such as network topology, profile information and user generated contents. Most of the previous researchers have focused on the structural features of the networks. While the recent researches indicate that contextual information can change the network topology. Although, there are number of valuable researches which combine structural and content information, but they face with the scalability issue due to feature engineering. Because, majority of the extracted features are obtained by a supervised or semi supervised algorithm. Moreover, the existing features are not general enough to indicate good performance on different networks with heterogeneous structures. Besides, most of the previous researches are presented for undirected and unweighted networks. In this paper, a novel link prediction framework called "DeepLink" is presented based on deep learning techniques. In contrast to the previous researches which fail to automatically extract best features for the link prediction, deep learning reduces the manual feature engineering. In this framework, both the structural and content information of the nodes are employed. The framework can use different structural feature vectors, which are prepared by various link prediction methods. It considers all proximity orders that are presented in a network during the structural feature learning. We have evaluated the performance of DeepLink on two real social network datasets including Telegram and irBlogs. On both datasets, the proposed framework outperforms several structural and hybrid approaches for link prediction problem.
Community Aware Random Walk for Network Embedding
Feb 19, 2018
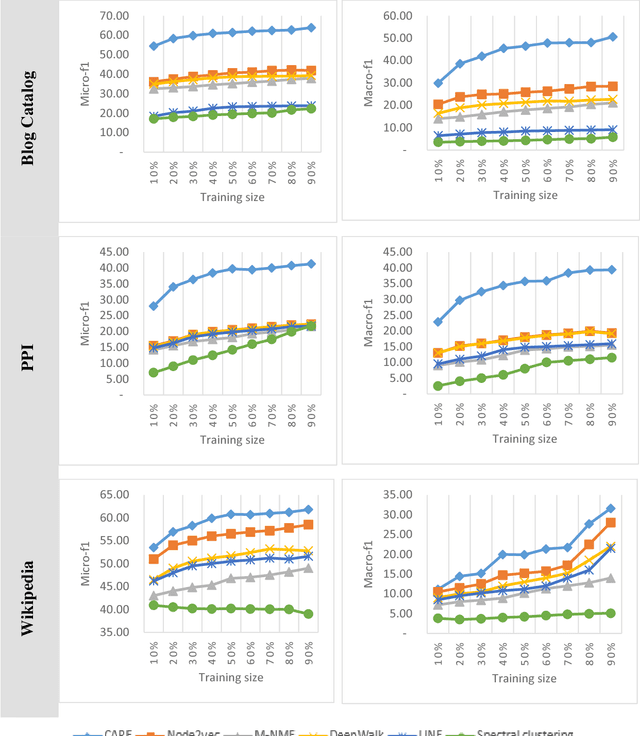
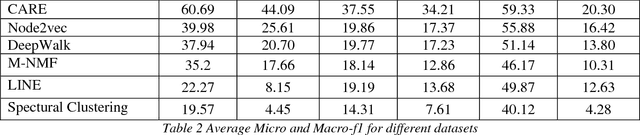
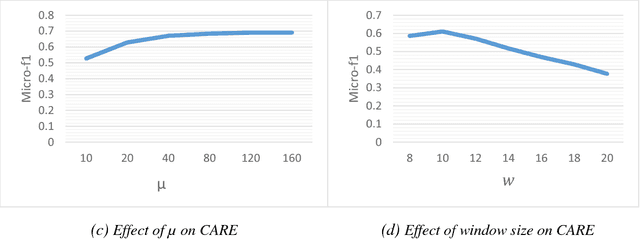
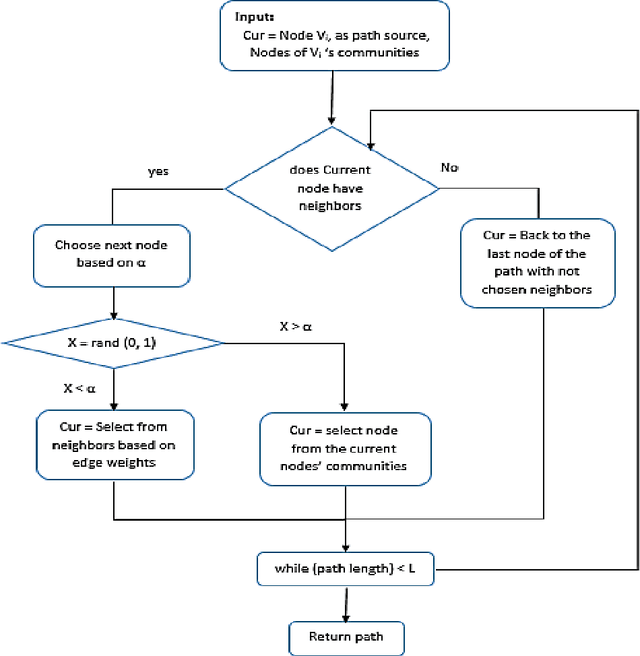
Abstract:Social network analysis provides meaningful information about behavior of network members that can be used for diverse applications such as classification, link prediction. However, network analysis is computationally expensive because of feature learning for different applications. In recent years, many researches have focused on feature learning methods in social networks. Network embedding represents the network in a lower dimensional representation space with the same properties which presents a compressed representation of the network. In this paper, we introduce a novel algorithm named "CARE" for network embedding that can be used for different types of networks including weighted, directed and complex. Current methods try to preserve local neighborhood information of nodes, whereas the proposed method utilizes local neighborhood and community information of network nodes to cover both local and global structure of social networks. CARE builds customized paths, which are consisted of local and global structure of network nodes, as a basis for network embedding and uses the Skip-gram model to learn representation vector of nodes. Subsequently, stochastic gradient descent is applied to optimize our objective function and learn the final representation of nodes. Our method can be scalable when new nodes are appended to network without information loss. Parallelize generation of customized random walks is also used for speeding up CARE. We evaluate the performance of CARE on multi label classification and link prediction tasks. Experimental results on various networks indicate that the proposed method outperforms others in both Micro and Macro-f1 measures for different size of training data.
* 17 pages, 3 figures, 4 Tables
Predicting protein-protein interactions based on rotation of proteins in 3D-space
Dec 22, 2017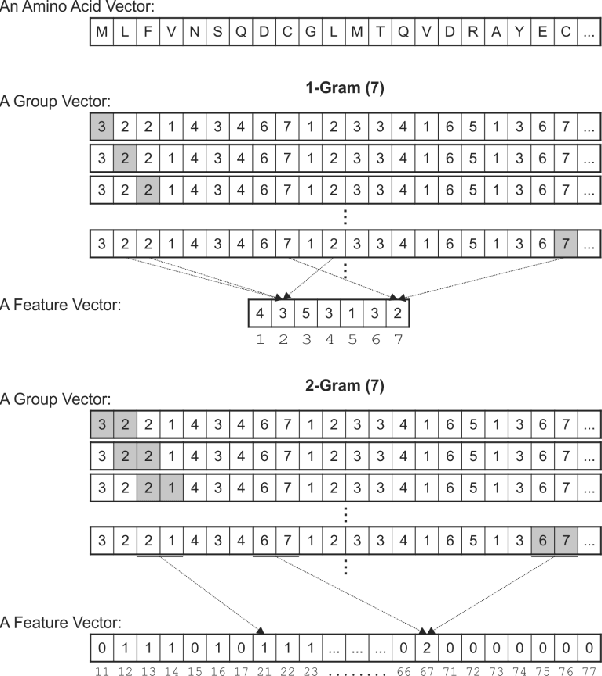
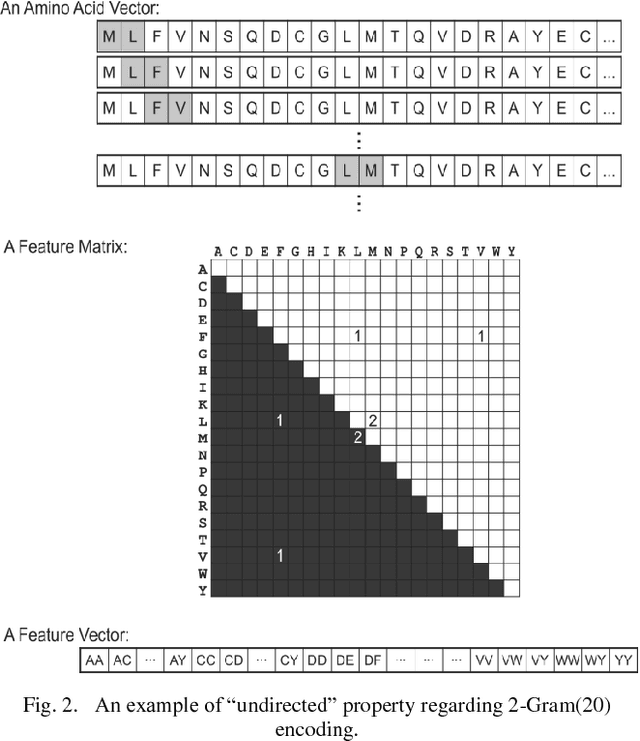
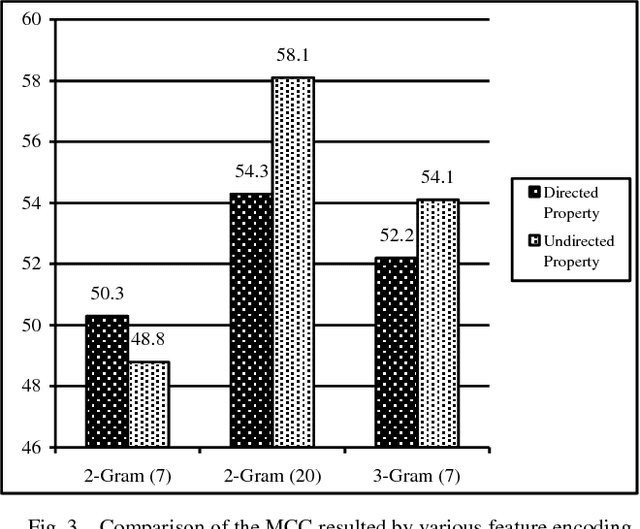
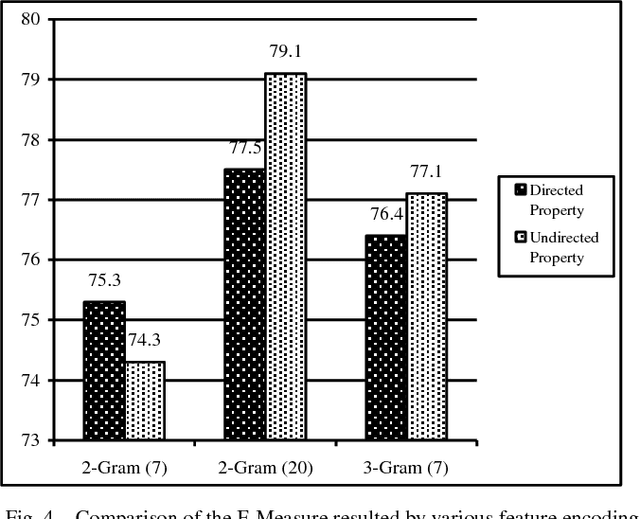
Abstract:Protein-Protein Interactions (PPIs) perform essential roles in biological functions. Although some experimental techniques have been developed to detect PPIs, they suffer from high false positive and high false negative rates. Consequently, efforts have been devoted during recent years to develop computational approaches to predict the interactions utilizing various sources of information. Therefore, a unique category of prediction approaches has been devised which is based on the protein sequence information. However, finding an appropriate feature encoding to characterize the sequence of proteins is a major challenge in such methods. In presented work, a sequence based method is proposed to predict protein-protein interactions using N-Gram encoding approaches to describe amino acids and a Relaxed Variable Kernel Density Estimator (RVKDE) as a machine learning tool. Moreover, since proteins can rotate in 3D-space, amino acid compositions have been considered with "undirected" property which leads to reduce dimensions of the vector space. The results show that our proposed method achieves the superiority of prediction performance with improving an F-measure of 2.5% on Human Protein Reference Dataset (HPRD).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge