Martin Hangaard Hansen
Autoencoding undirected molecular graphs with neural networks
Nov 26, 2019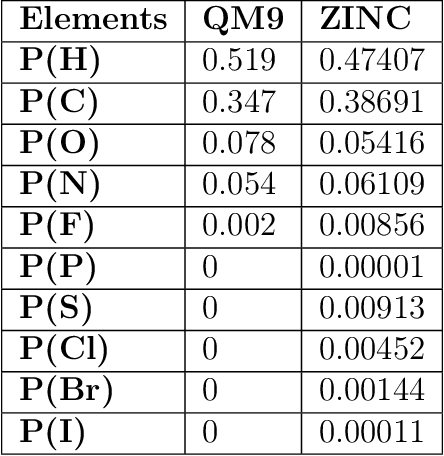
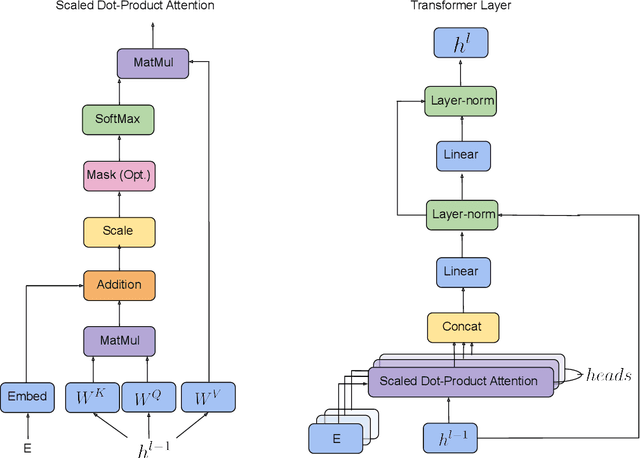

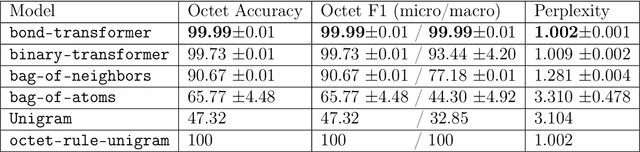
Abstract:We propose a machine learning model, inspired by language modeling from natural language processing, which can automatically correct molecules in discrete representations using a structure rule learned from a collection of undirected molecular graphs. Using discrete representations of molecules allows cheap, fast, and coarse grained insights. We introduce an adaption on a modern neural network architecture, the Transformer, which can learn relationships between atoms and bonds. The algorithm thereby solves the unsupervised task of recovering partially observed molecules represented as undirected graphs. This is to our knowledge, the first work that can automatically learn any discrete molecular structure rule with input exclusively consisting of a training set of molecules. In this work the neural network successfully approximates the octet rule, relations in hypervalent molecules and ions when trained on the ZINC and QM9 dataset. These results provides encouraging evidence that neural networks can learn advanced molecular structure rules and dataset specific properties, as the transformer surpasses a strong octet-rule baseline.
An Atomistic Machine Learning Package for Surface Science and Catalysis
Apr 01, 2019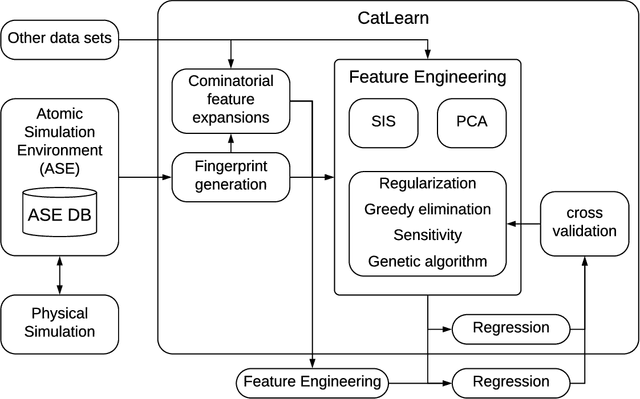
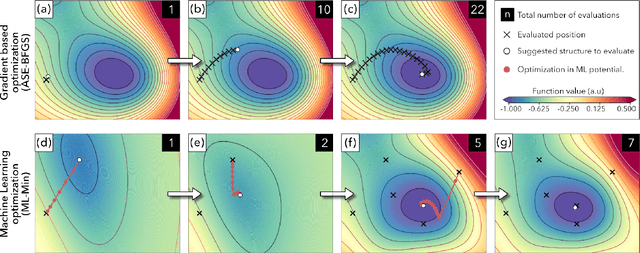
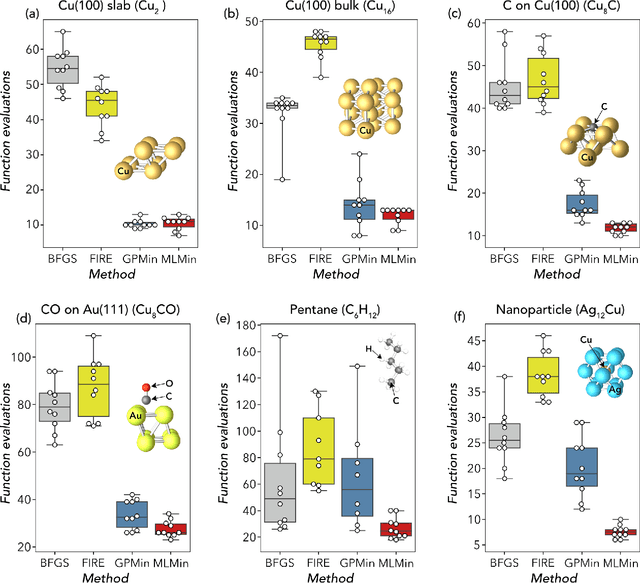
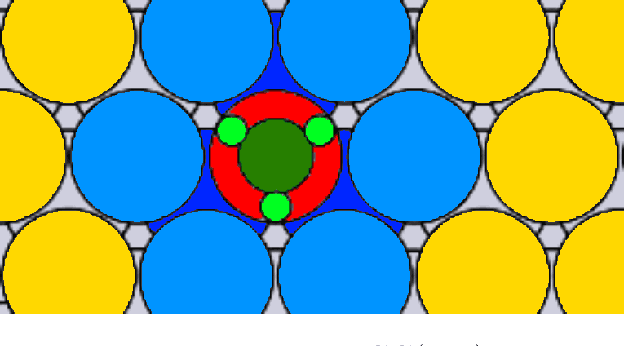
Abstract:We present work flows and a software module for machine learning model building in surface science and heterogeneous catalysis. This includes fingerprinting atomic structures from 3D structure and/or connectivity information, it includes descriptor selection methods and benchmarks, and it includes active learning frameworks for atomic structure optimization, acceleration of screening studies and for exploration of the structure space of nano particles, which are all atomic structure problems relevant for surface science and heterogeneous catalysis. Our overall goal is to provide a repository to ease machine learning model building for catalysis, to advance the models beyond the chemical intuition of the user and to increase autonomy for exploration of chemical space.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge