Mark C. Walker
Automated Classification of First-Trimester Fetal Heart Views Using Ultrasound-Specific Self-Supervised Learning
Dec 30, 2025Abstract:Congenital heart disease remains the most common congenital anomaly and a leading cause of neonatal morbidity and mortality. Although first-trimester fetal echocardiography offers an opportunity for earlier detection, automated analysis at this stage is challenging due to small cardiac structures, low signal-to-noise ratio, and substantial inter-operator variability. In this work, we evaluate a self-supervised ultrasound foundation model, USF-MAE, for first-trimester fetal heart view classification. USF-MAE is pretrained using masked autoencoding modelling on more than 370,000 unlabelled ultrasound images spanning over 40 anatomical regions and is subsequently fine-tuned for downstream classification. As a proof of concept, the pretrained Vision Transformer encoder was fine-tuned on an open-source dataset of 6,720 first-trimester fetal echocardiography images to classify five categories: aorta, atrioventricular flows, V sign, X sign, and Other. Model performance was benchmarked against supervised convolutional neural network baselines (ResNet-18 and ResNet-50) and a Vision Transformer (ViT-B/16) model pretrained on natural images (ImageNet-1k). All models were trained and evaluated using identical preprocessing, data splits, and optimization protocols. On an independent test set, USF-MAE achieved the highest performance across all evaluation metrics, with 90.57% accuracy, 91.15% precision, 90.57% recall, and 90.71% F1-score. This represents an improvement of +2.03% in accuracy and +1.98% in F1-score compared with the strongest baseline, ResNet-18. The proposed approach demonstrated robust performance without reliance on aggressive image preprocessing or region-of-interest cropping and showed improved discrimination of non-diagnostic frames.
Self-Supervised Ultrasound Representation Learning for Renal Anomaly Prediction in Prenatal Imaging
Dec 15, 2025



Abstract:Prenatal ultrasound is the cornerstone for detecting congenital anomalies of the kidneys and urinary tract, but diagnosis is limited by operator dependence and suboptimal imaging conditions. We sought to assess the performance of a self-supervised ultrasound foundation model for automated fetal renal anomaly classification using a curated dataset of 969 two-dimensional ultrasound images. A pretrained Ultrasound Self-Supervised Foundation Model with Masked Autoencoding (USF-MAE) was fine-tuned for binary and multi-class classification of normal kidneys, urinary tract dilation, and multicystic dysplastic kidney. Models were compared with a DenseNet-169 convolutional baseline using cross-validation and an independent test set. USF-MAE consistently improved upon the baseline across all evaluation metrics in both binary and multi-class settings. USF-MAE achieved an improvement of about 1.87% (AUC) and 7.8% (F1-score) on the validation set, 2.32% (AUC) and 4.33% (F1-score) on the independent holdout test set. The largest gains were observed in the multi-class setting, where the improvement in AUC was 16.28% and 46.15% in F1-score. To facilitate model interpretability, Score-CAM visualizations were adapted for a transformer architecture and show that model predictions were informed by known, clinically relevant renal structures, including the renal pelvis in urinary tract dilation and cystic regions in multicystic dysplastic kidney. These results show that ultrasound-specific self-supervised learning can generate a useful representation as a foundation for downstream diagnostic tasks. The proposed framework offers a robust, interpretable approach to support the prenatal detection of renal anomalies and demonstrates the promise of foundation models in obstetric imaging.
CAManim: Animating end-to-end network activation maps
Dec 19, 2023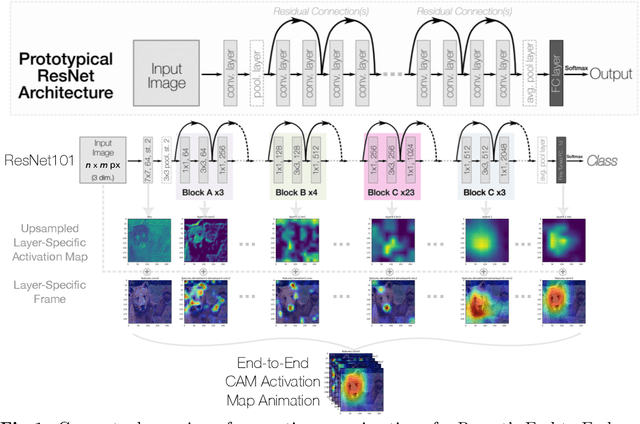
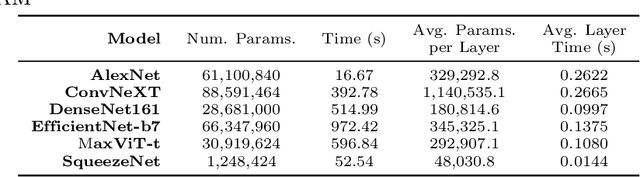
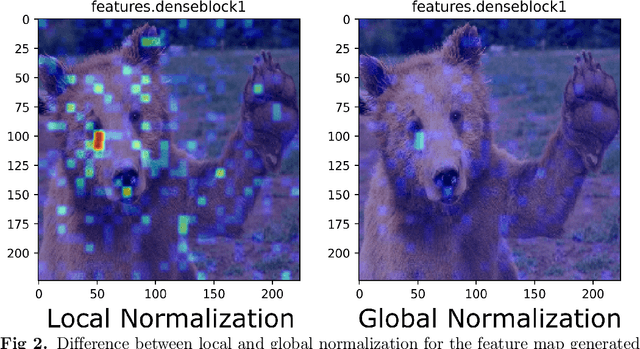
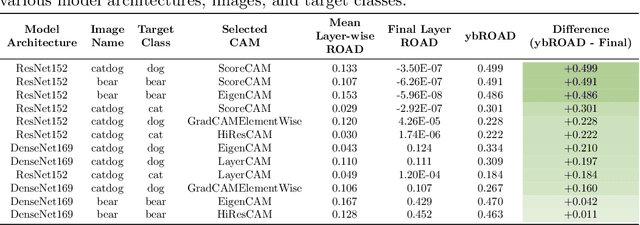
Abstract:Deep neural networks have been widely adopted in numerous domains due to their high performance and accessibility to developers and application-specific end-users. Fundamental to image-based applications is the development of Convolutional Neural Networks (CNNs), which possess the ability to automatically extract features from data. However, comprehending these complex models and their learned representations, which typically comprise millions of parameters and numerous layers, remains a challenge for both developers and end-users. This challenge arises due to the absence of interpretable and transparent tools to make sense of black-box models. There exists a growing body of Explainable Artificial Intelligence (XAI) literature, including a collection of methods denoted Class Activation Maps (CAMs), that seek to demystify what representations the model learns from the data, how it informs a given prediction, and why it, at times, performs poorly in certain tasks. We propose a novel XAI visualization method denoted CAManim that seeks to simultaneously broaden and focus end-user understanding of CNN predictions by animating the CAM-based network activation maps through all layers, effectively depicting from end-to-end how a model progressively arrives at the final layer activation. Herein, we demonstrate that CAManim works with any CAM-based method and various CNN architectures. Beyond qualitative model assessments, we additionally propose a novel quantitative assessment that expands upon the Remove and Debias (ROAD) metric, pairing the qualitative end-to-end network visual explanations assessment with our novel quantitative "yellow brick ROAD" assessment (ybROAD). This builds upon prior research to address the increasing demand for interpretable, robust, and transparent model assessment methodology, ultimately improving an end-user's trust in a given model's predictions.
MetaCAM: Ensemble-Based Class Activation Map
Jul 31, 2023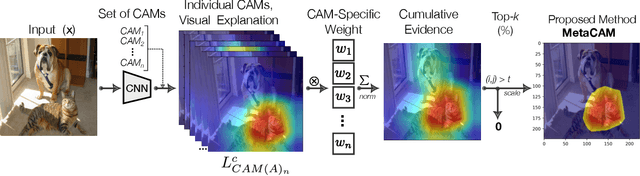
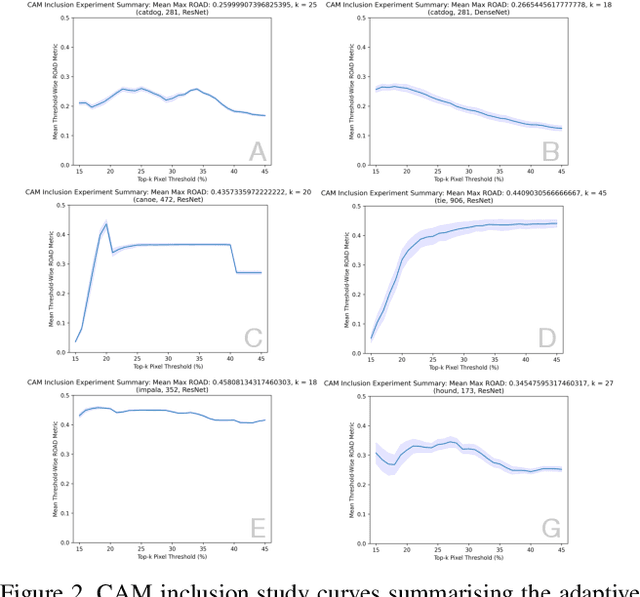
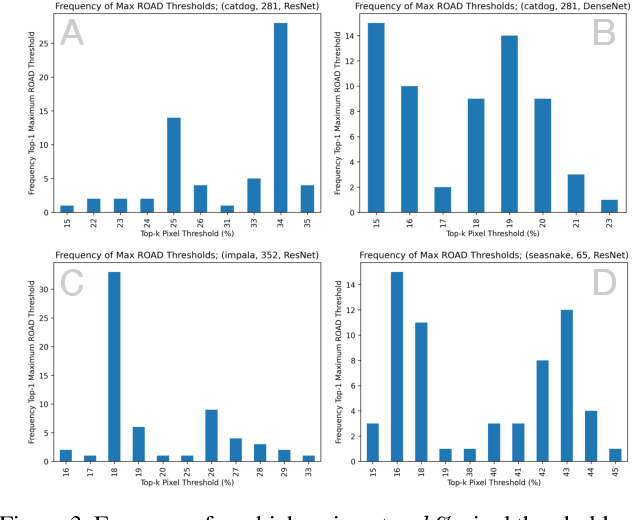
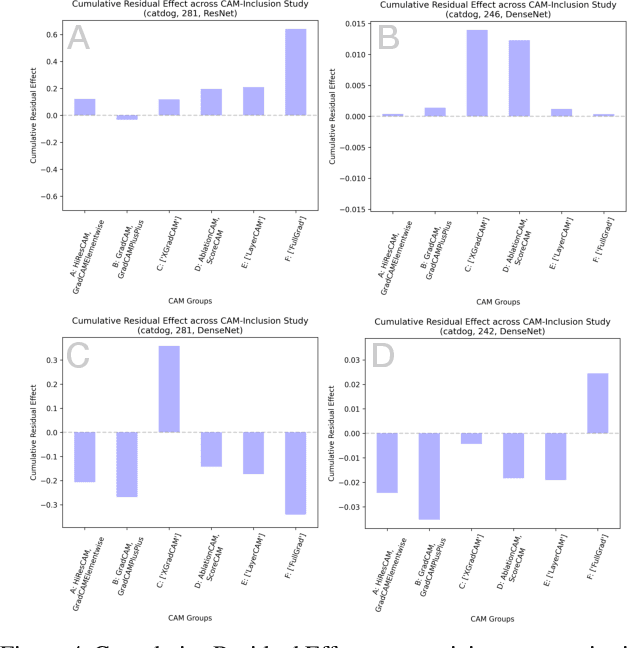
Abstract:The need for clear, trustworthy explanations of deep learning model predictions is essential for high-criticality fields, such as medicine and biometric identification. Class Activation Maps (CAMs) are an increasingly popular category of visual explanation methods for Convolutional Neural Networks (CNNs). However, the performance of individual CAMs depends largely on experimental parameters such as the selected image, target class, and model. Here, we propose MetaCAM, an ensemble-based method for combining multiple existing CAM methods based on the consensus of the top-k% most highly activated pixels across component CAMs. We perform experiments to quantifiably determine the optimal combination of 11 CAMs for a given MetaCAM experiment. A new method denoted Cumulative Residual Effect (CRE) is proposed to summarize large-scale ensemble-based experiments. We also present adaptive thresholding and demonstrate how it can be applied to individual CAMs to improve their performance, measured using pixel perturbation method Remove and Debias (ROAD). Lastly, we show that MetaCAM outperforms existing CAMs and refines the most salient regions of images used for model predictions. In a specific example, MetaCAM improved ROAD performance to 0.393 compared to 11 individual CAMs with ranges from -0.101-0.172, demonstrating the importance of combining CAMs through an ensembling method and adaptive thresholding.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge