Marco Frasca
Quantization-Aware Regularizers for Deep Neural Networks Compression
Feb 03, 2026Abstract:Deep Neural Networks reached state-of-the-art performance across numerous domains, but this progress has come at the cost of increasingly large and over-parameterized models, posing serious challenges for deployment on resource-constrained devices. As a result, model compression has become essential, and -- among compression techniques -- weight quantization is largely used and particularly effective, yet it typically introduces a non-negligible accuracy drop. However, it is usually applied to already trained models, without influencing how the parameter space is explored during the learning phase. In contrast, we introduce per-layer regularization terms that drive weights to naturally form clusters during training, integrating quantization awareness directly into the optimization process. This reduces the accuracy loss typically associated with quantization methods while preserving their compression potential. Furthermore, in our framework quantization representatives become network parameters, marking, to the best of our knowledge, the first approach to embed quantization parameters directly into the backpropagation procedure. Experiments on CIFAR-10 with AlexNet and VGG16 models confirm the effectiveness of the proposed strategy.
A Critical Analysis of Classifier Selection in Learned Bloom Filters
Nov 28, 2022Abstract:Learned Bloom Filters, i.e., models induced from data via machine learning techniques and solving the approximate set membership problem, have recently been introduced with the aim of enhancing the performance of standard Bloom Filters, with special focus on space occupancy. Unlike in the classical case, the "complexity" of the data used to build the filter might heavily impact on its performance. Therefore, here we propose the first in-depth analysis, to the best of our knowledge, for the performance assessment of a given Learned Bloom Filter, in conjunction with a given classifier, on a dataset of a given classification complexity. Indeed, we propose a novel methodology, supported by software, for designing, analyzing and implementing Learned Bloom Filters in function of specific constraints on their multi-criteria nature (that is, constraints involving space efficiency, false positive rate, and reject time). Our experiments show that the proposed methodology and the supporting software are valid and useful: we find out that only two classifiers have desirable properties in relation to problems with different data complexity, and, interestingly, none of them has been considered so far in the literature. We also experimentally show that the Sandwiched variant of Learned Bloom filters is the most robust to data complexity and classifier performance variability, as well as those usually having smaller reject times. The software can be readily used to test new Learned Bloom Filter proposals, which can be compared with the best ones identified here.
On the Choice of General Purpose Classifiers in Learned Bloom Filters: An Initial Analysis Within Basic Filters
Dec 13, 2021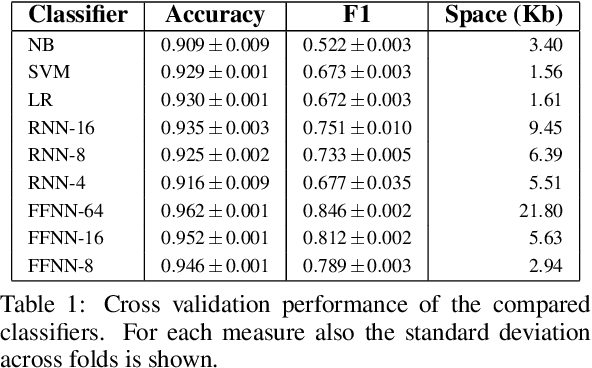
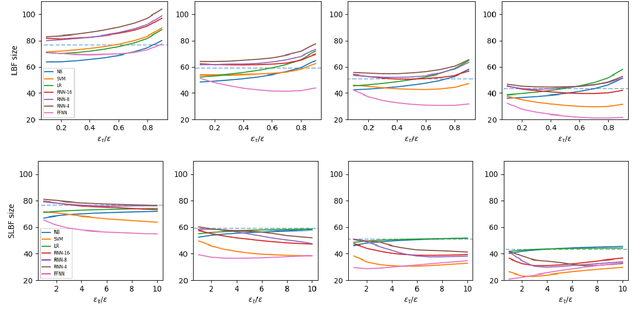
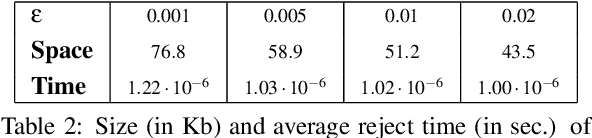
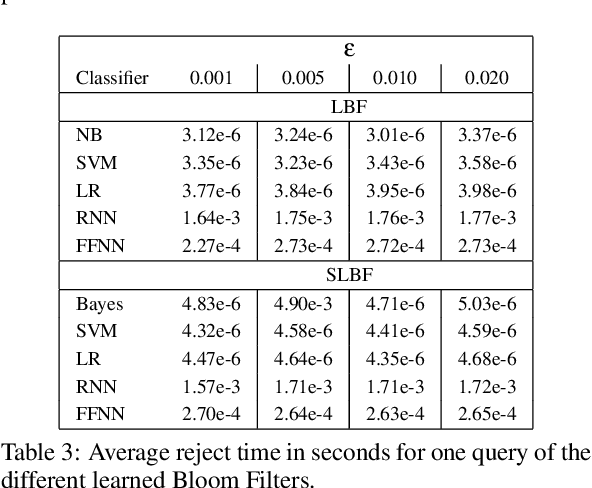
Abstract:Bloom Filters are a fundamental and pervasive data structure. Within the growing area of Learned Data Structures, several Learned versions of Bloom Filters have been considered, yielding advantages over classic Filters. Each of them uses a classifier, which is the Learned part of the data structure. Although it has a central role in those new filters, and its space footprint as well as classification time may affect the performance of the Learned Filter, no systematic study of which specific classifier to use in which circumstances is available. We report progress in this area here, providing also initial guidelines on which classifier to choose among five classic classification paradigms.
Compact representations of convolutional neural networks via weight pruning and quantization
Aug 28, 2021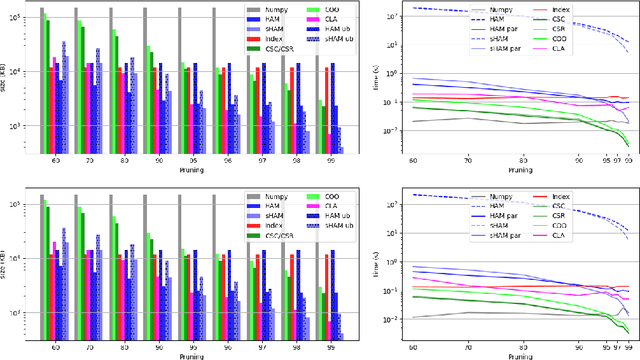
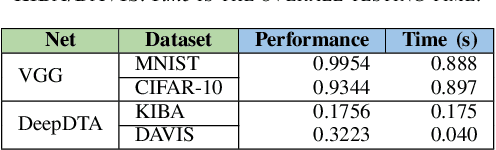
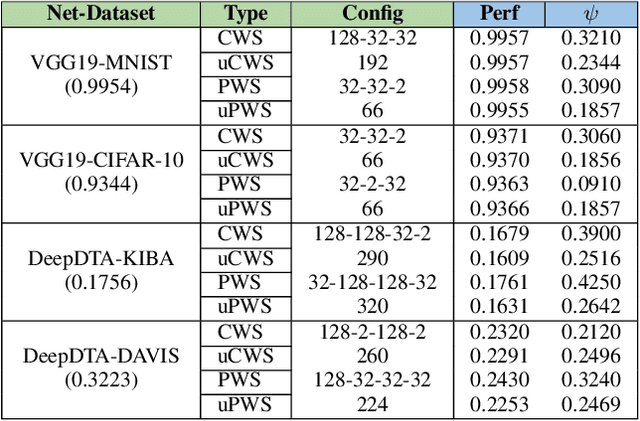
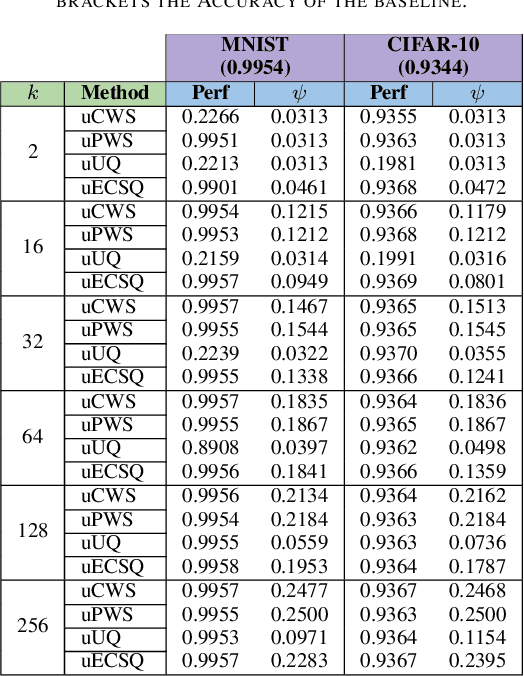
Abstract:The state-of-the-art performance for several real-world problems is currently reached by convolutional neural networks (CNN). Such learning models exploit recent results in the field of deep learning, typically leading to highly performing, yet very large neural networks with (at least) millions of parameters. As a result, the deployment of such models is not possible when only small amounts of RAM are available, or in general within resource-limited platforms, and strategies to compress CNNs became thus of paramount importance. In this paper we propose a novel lossless storage format for CNNs based on source coding and leveraging both weight pruning and quantization. We theoretically derive the space upper bounds for the proposed structures, showing their relationship with both sparsity and quantization levels of the weight matrices. Both compression rates and excution times have been tested against reference methods for matrix compression, and an empirical evaluation of state-of-the-art quantization schemes based on weight sharing is also discussed, to assess their impact on the performance when applied to both convolutional and fully connected layers. On four benchmarks for classification and regression problems and comparing to the baseline pre-trained uncompressed network, we achieved a reduction of space occupancy up to 0.6% on fully connected layers and 5.44% on the whole network, while performing at least as competitive as the baseline.
Compression strategies and space-conscious representations for deep neural networks
Jul 15, 2020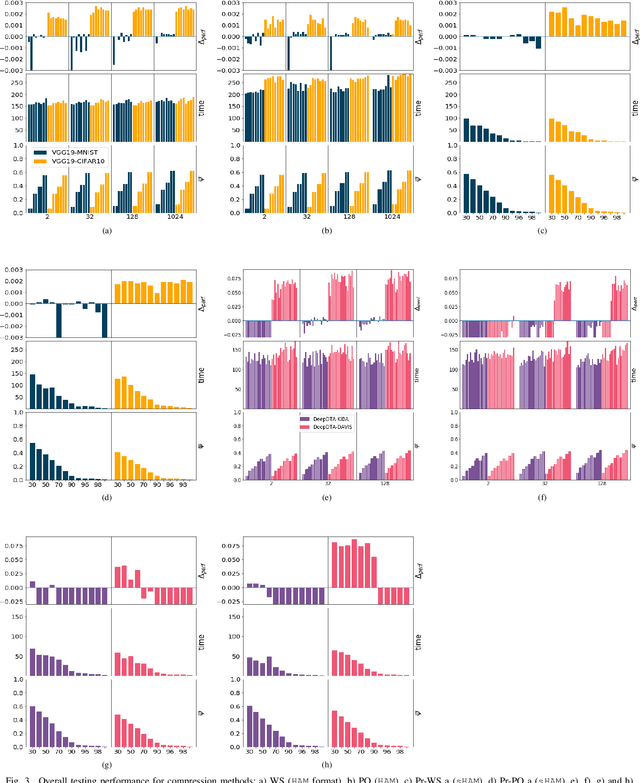
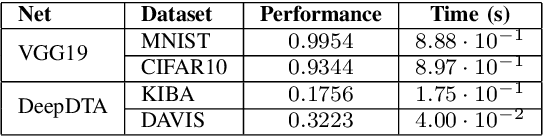
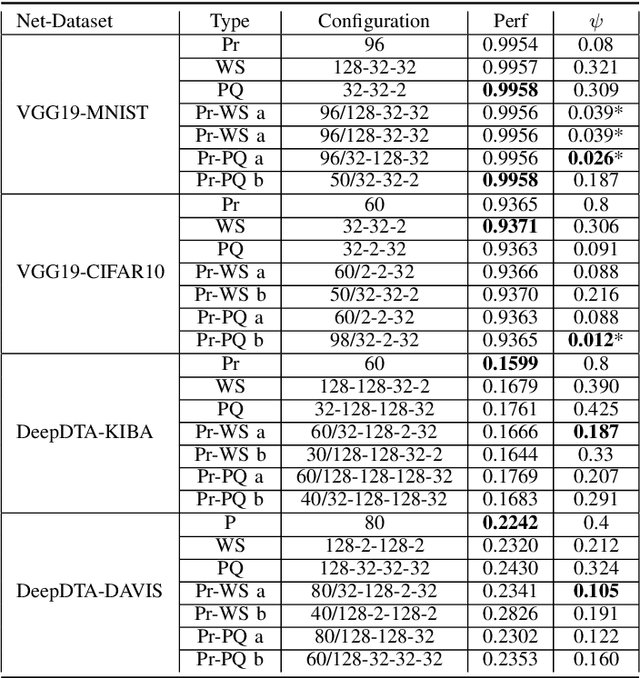
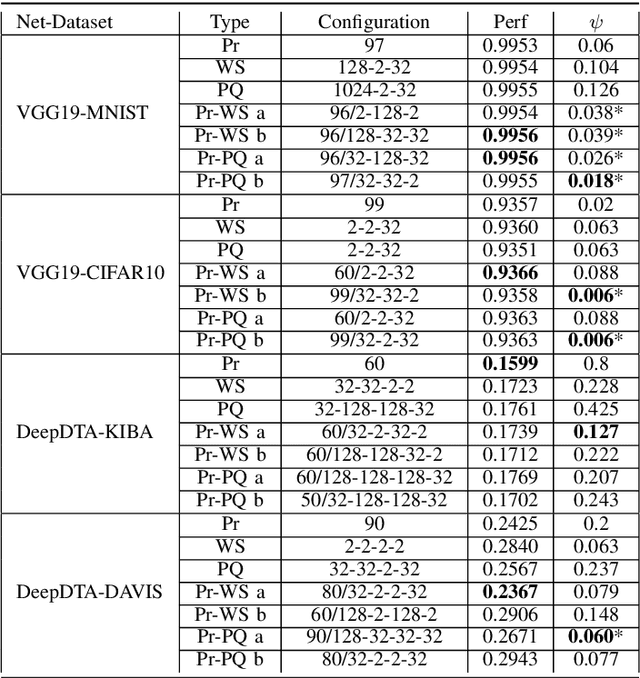
Abstract:Recent advances in deep learning have made available large, powerful convolutional neural networks (CNN) with state-of-the-art performance in several real-world applications. Unfortunately, these large-sized models have millions of parameters, thus they are not deployable on resource-limited platforms (e.g. where RAM is limited). Compression of CNNs thereby becomes a critical problem to achieve memory-efficient and possibly computationally faster model representations. In this paper, we investigate the impact of lossy compression of CNNs by weight pruning and quantization, and lossless weight matrix representations based on source coding. We tested several combinations of these techniques on four benchmark datasets for classification and regression problems, achieving compression rates up to $165$ times, while preserving or improving the model performance.
Multitask Hopfield Networks
Apr 10, 2019
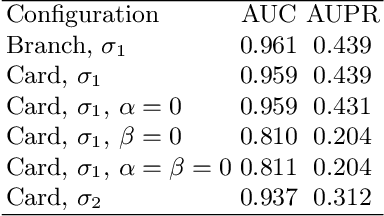
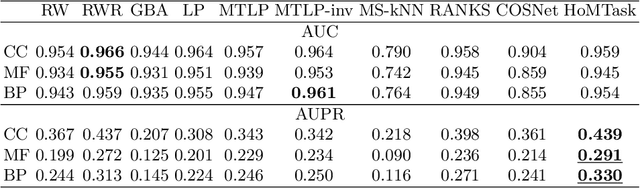
Abstract:Multitask algorithms typically use task similarity information as a bias to speed up and improve the performance of learning processes. Tasks are learned jointly, sharing information across them, in order to construct models more accurate than those learned separately over single tasks. In this contribution, we present the first multitask model, to our knowledge, based on Hopfield Networks (HNs), named HoMTask. We show that by appropriately building a unique HN embedding all tasks, a more robust and effective classification model can be learned. HoMTask is a transductive semi-supervised parametric HN, that minimizes an energy function extended to all nodes and to all tasks under study. We provide theoretical evidence that the optimal parameters automatically estimated by HoMTask make coherent the model itself with the prior knowledge (connection weights and node labels). The convergence properties of HNs are preserved, and the fixed point reached by the network dynamics gives rise to the prediction of unlabeled nodes. The proposed model improves the classification abilities of singletask HNs on a preliminary benchmark comparison, and achieves competitive performance with state-of-the-art semi-supervised graph-based algorithms.
Combining Cost-Sensitive Classification with Negative Selection for Protein Function Prediction
May 18, 2018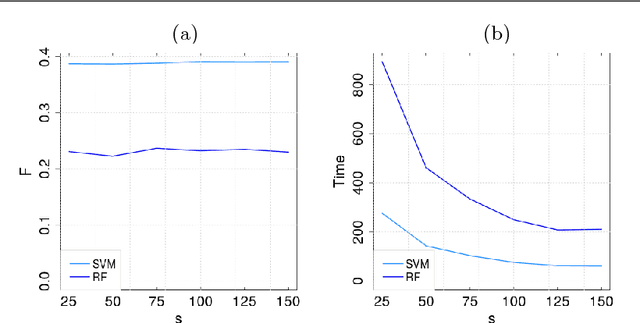
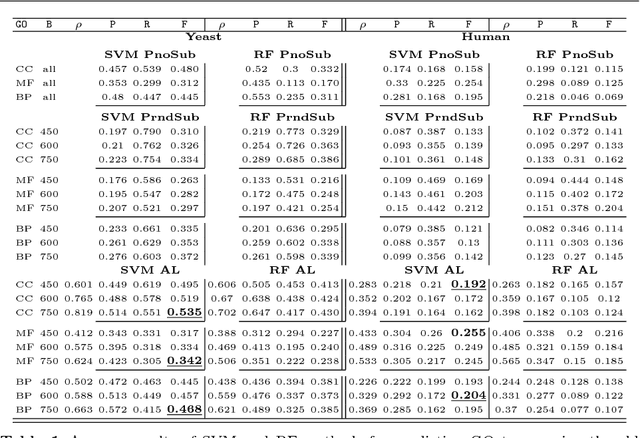
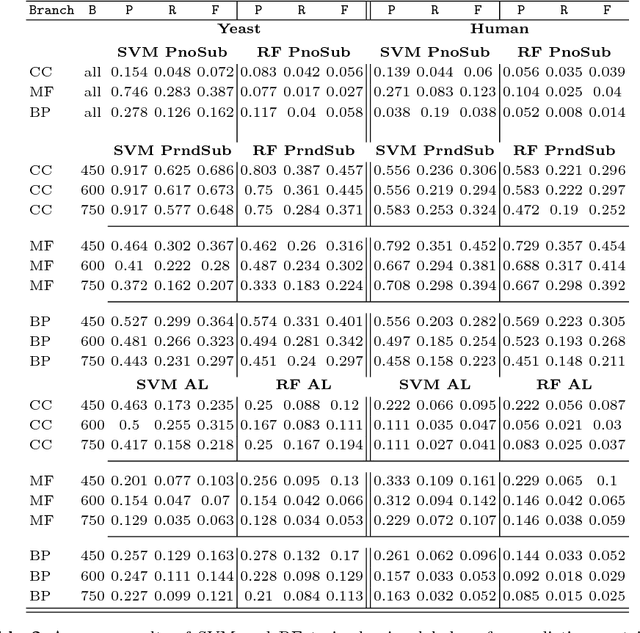
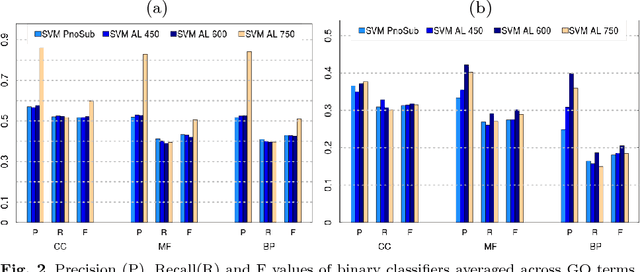
Abstract:Motivation: Computational methods play a central role in annotating the functions of large amounts of proteins delivered by high-throughput technologies. Despite the encouraging results achieved by these methods, many functions still have a very low number of verified protein annotations, leading to a pronounced imbalance between annotated and unannotated proteins. Furthermore, functional taxonomies rarely report negative annotations. This leaves ill defined the set of negative examples, which is crucial for training the majority of machine learning methods. In practice, neglecting data imbalance and the problem of selecing negative examples can strongly limit the accuracy of protein function prediction. Results: We present a novel approach combining a suitable imbalance-aware classification strategy, addressing the scarcity of annotated proteins, with an active learning strategy for selecting the most reliable negative examples. When implemented in a Support Vector Machine, this combined approach shows improved accuracy on yeast and human proteomes over standard SVM and top-performing function prediction tools
Multitask Protein Function Prediction Through Task Dissimilarity
Nov 03, 2016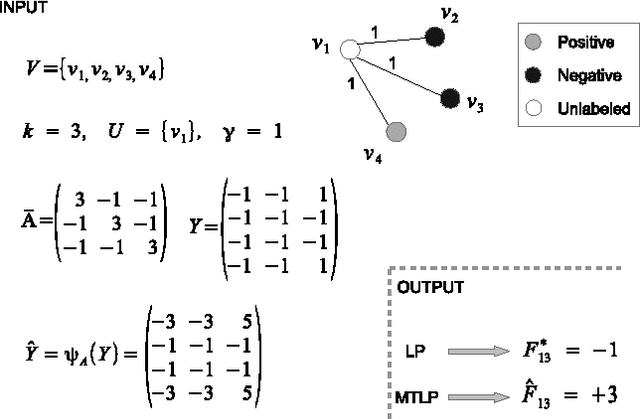
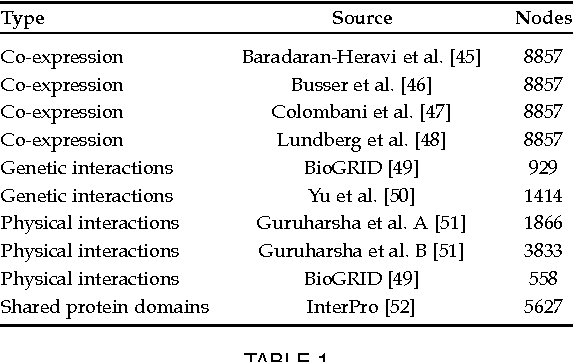
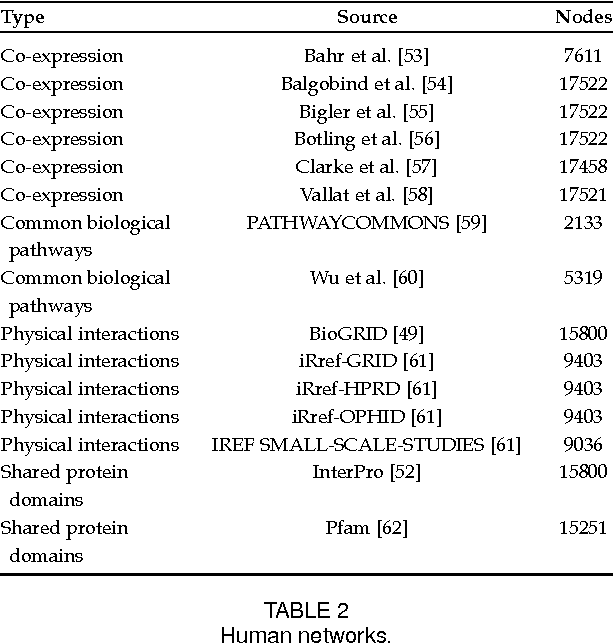

Abstract:Automated protein function prediction is a challenging problem with distinctive features, such as the hierarchical organization of protein functions and the scarcity of annotated proteins for most biological functions. We propose a multitask learning algorithm addressing both issues. Unlike standard multitask algorithms, which use task (protein functions) similarity information as a bias to speed up learning, we show that dissimilarity information enforces separation of rare class labels from frequent class labels, and for this reason is better suited for solving unbalanced protein function prediction problems. We support our claim by showing that a multitask extension of the label propagation algorithm empirically works best when the task relatedness information is represented using a dissimilarity matrix as opposed to a similarity matrix. Moreover, the experimental comparison carried out on three model organism shows that our method has a more stable performance in both "protein-centric" and "function-centric" evaluation settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge