Luqin Gan
Are machine learning interpretations reliable? A stability study on global interpretations
May 21, 2025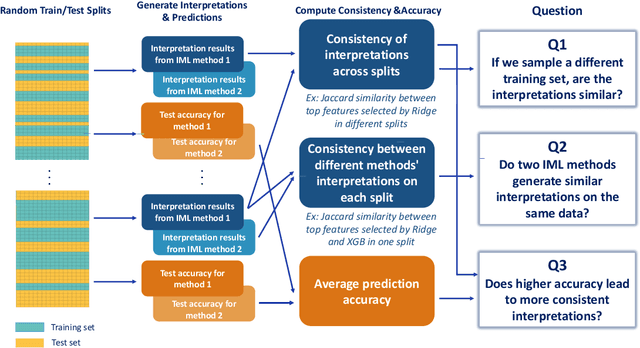
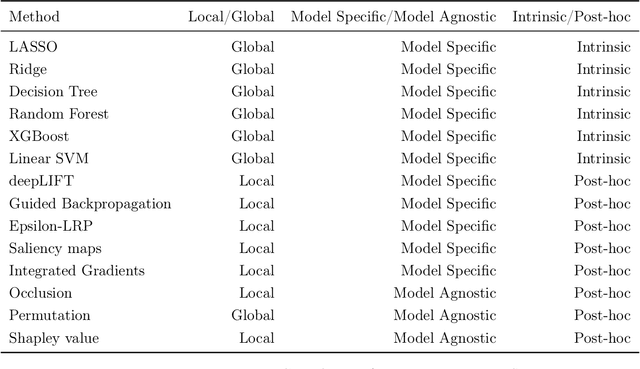
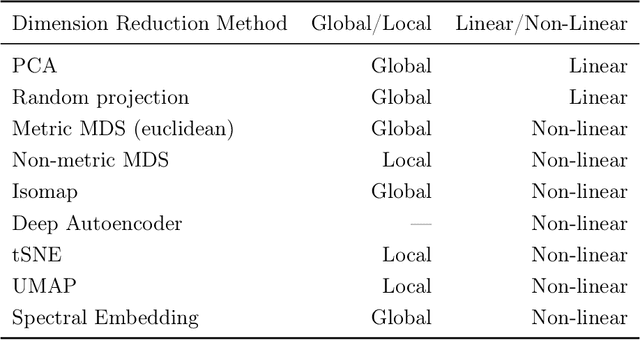
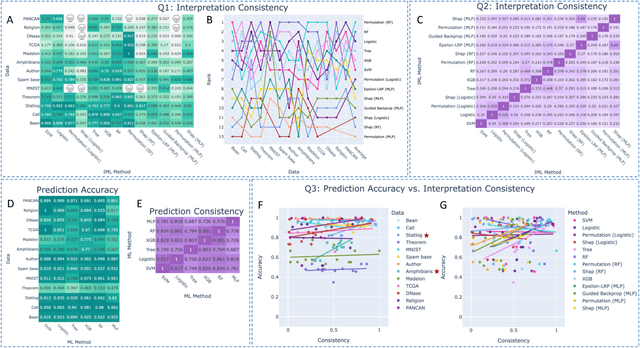
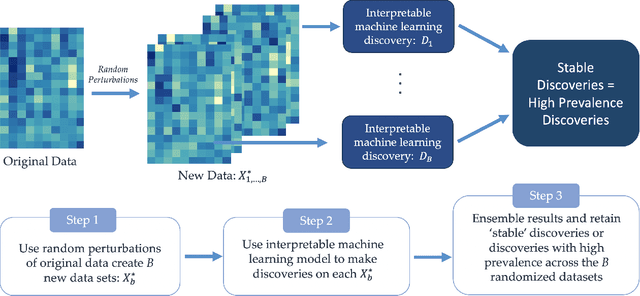
Abstract:As machine learning systems are increasingly used in high-stakes domains, there is a growing emphasis placed on making them interpretable to improve trust in these systems. In response, a range of interpretable machine learning (IML) methods have been developed to generate human-understandable insights into otherwise black box models. With these methods, a fundamental question arises: Are these interpretations reliable? Unlike with prediction accuracy or other evaluation metrics for supervised models, the proximity to the true interpretation is difficult to define. Instead, we ask a closely related question that we argue is a prerequisite for reliability: Are these interpretations stable? We define stability as findings that are consistent or reliable under small random perturbations to the data or algorithms. In this study, we conduct the first systematic, large-scale empirical stability study on popular machine learning global interpretations for both supervised and unsupervised tasks on tabular data. Our findings reveal that popular interpretation methods are frequently unstable, notably less stable than the predictions themselves, and that there is no association between the accuracy of machine learning predictions and the stability of their associated interpretations. Moreover, we show that no single method consistently provides the most stable interpretations across a range of benchmark datasets. Overall, these results suggest that interpretability alone does not warrant trust, and underscores the need for rigorous evaluation of interpretation stability in future work. To support these principles, we have developed and released an open source IML dashboard and Python package to enable researchers to assess the stability and reliability of their own data-driven interpretations and discoveries.
Interpretable Machine Learning for Discovery: Statistical Challenges \& Opportunities
Aug 02, 2023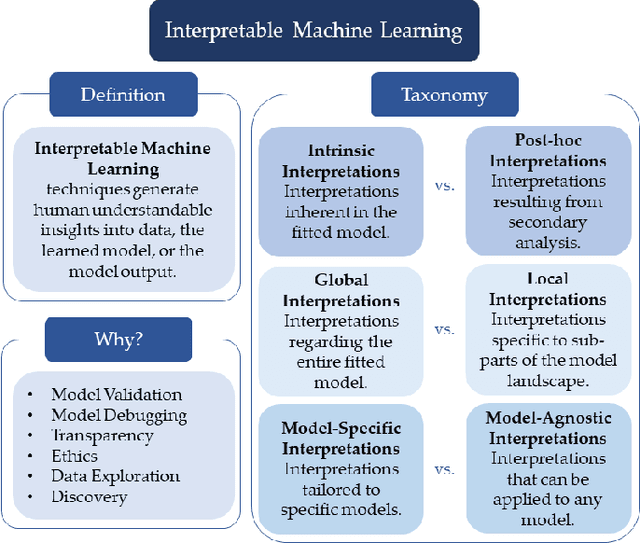
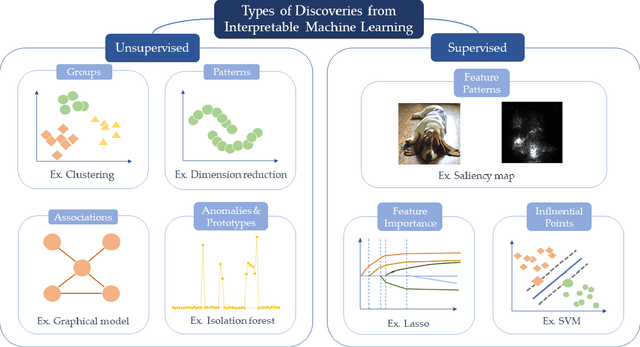
Abstract:New technologies have led to vast troves of large and complex datasets across many scientific domains and industries. People routinely use machine learning techniques to not only process, visualize, and make predictions from this big data, but also to make data-driven discoveries. These discoveries are often made using Interpretable Machine Learning, or machine learning models and techniques that yield human understandable insights. In this paper, we discuss and review the field of interpretable machine learning, focusing especially on the techniques as they are often employed to generate new knowledge or make discoveries from large data sets. We outline the types of discoveries that can be made using Interpretable Machine Learning in both supervised and unsupervised settings. Additionally, we focus on the grand challenge of how to validate these discoveries in a data-driven manner, which promotes trust in machine learning systems and reproducibility in science. We discuss validation from both a practical perspective, reviewing approaches based on data-splitting and stability, as well as from a theoretical perspective, reviewing statistical results on model selection consistency and uncertainty quantification via statistical inference. Finally, we conclude by highlighting open challenges in using interpretable machine learning techniques to make discoveries, including gaps between theory and practice for validating data-driven-discoveries.
Inference for Interpretable Machine Learning: Fast, Model-Agnostic Confidence Intervals for Feature Importance
Jun 05, 2022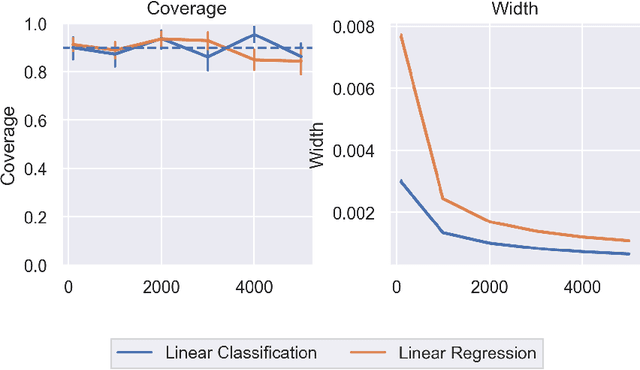
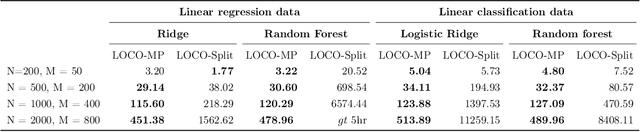
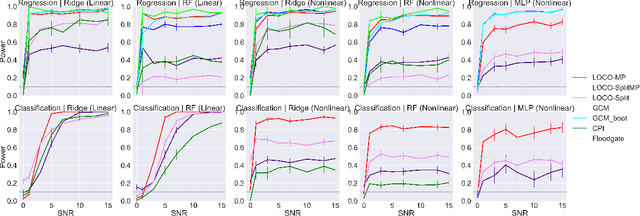
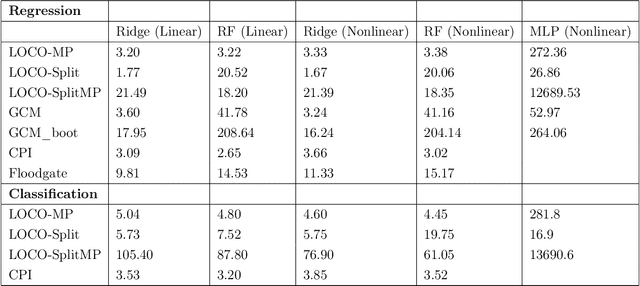
Abstract:In order to trust machine learning for high-stakes problems, we need models to be both reliable and interpretable. Recently, there has been a growing body of work on interpretable machine learning which generates human understandable insights into data, models, or predictions. At the same time, there has been increased interest in quantifying the reliability and uncertainty of machine learning predictions, often in the form of confidence intervals for predictions using conformal inference. Yet, there has been relatively little attention given to the reliability and uncertainty of machine learning interpretations, which is the focus of this paper. Our goal is to develop confidence intervals for a widely-used form of machine learning interpretation: feature importance. We specifically seek to develop universal model-agnostic and assumption-light confidence intervals for feature importance that will be valid for any machine learning model and for any regression or classification task. We do so by leveraging a form of random observation and feature subsampling called minipatch ensembles and show that our approach provides assumption-light asymptotic coverage for the feature importance score of any model. Further, our approach is fast as computations needed for inference come nearly for free as part of the ensemble learning process. Finally, we also show that our same procedure can be leveraged to provide valid confidence intervals for predictions, hence providing fast, simultaneous quantification of the uncertainty of both model predictions and interpretations. We validate our intervals on a series of synthetic and real data examples, showing that our approach detects the correct important features and exhibits many computational and statistical advantages over existing methods.
Fast and Interpretable Consensus Clustering via Minipatch Learning
Oct 18, 2021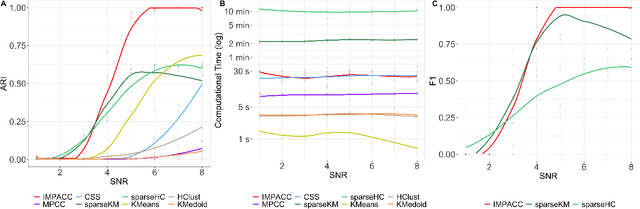
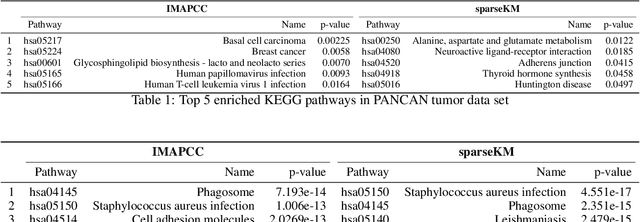
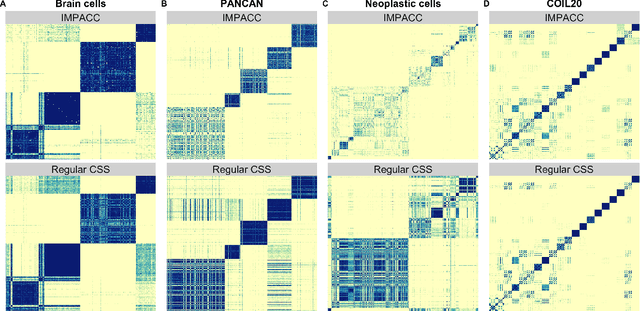
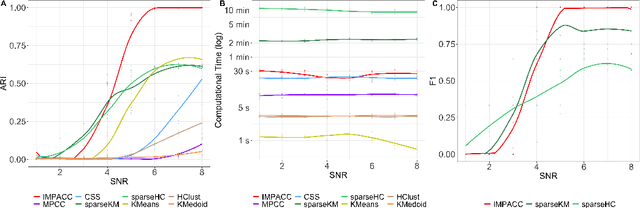
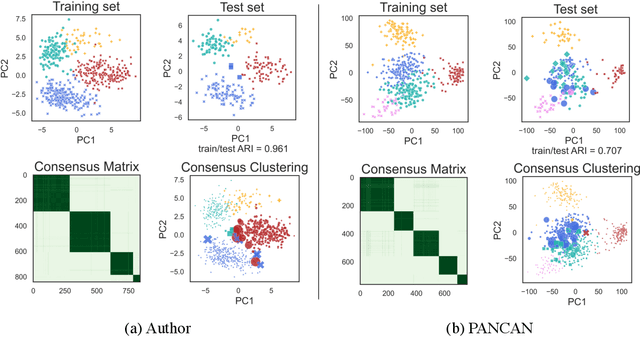
Abstract:Consensus clustering has been widely used in bioinformatics and other applications to improve the accuracy, stability and reliability of clustering results. This approach ensembles cluster co-occurrences from multiple clustering runs on subsampled observations. For application to large-scale bioinformatics data, such as to discover cell types from single-cell sequencing data, for example, consensus clustering has two significant drawbacks: (i) computational inefficiency due to repeatedly applying clustering algorithms, and (ii) lack of interpretability into the important features for differentiating clusters. In this paper, we address these two challenges by developing IMPACC: Interpretable MiniPatch Adaptive Consensus Clustering. Our approach adopts three major innovations. We ensemble cluster co-occurrences from tiny subsets of both observations and features, termed minipatches, thus dramatically reducing computation time. Additionally, we develop adaptive sampling schemes for observations, which result in both improved reliability and computational savings, as well as adaptive sampling schemes of features, which leads to interpretable solutions by quickly learning the most relevant features that differentiate clusters. We study our approach on synthetic data and a variety of real large-scale bioinformatics data sets; results show that our approach not only yields more accurate and interpretable cluster solutions, but it also substantially improves computational efficiency compared to standard consensus clustering approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge