Luis Rueda
Vectorized Context-Aware Embeddings for GAT-Based Collaborative Filtering
Oct 30, 2025Abstract:Recommender systems often struggle with data sparsity and cold-start scenarios, limiting their ability to provide accurate suggestions for new or infrequent users. This paper presents a Graph Attention Network (GAT) based Collaborative Filtering (CF) framework enhanced with Large Language Model (LLM) driven context aware embeddings. Specifically, we generate concise textual user profiles and unify item metadata (titles, genres, overviews) into rich textual embeddings, injecting these as initial node features in a bipartite user item graph. To further optimize ranking performance, we introduce a hybrid loss function that combines Bayesian Personalized Ranking (BPR) with a cosine similarity term and robust negative sampling, ensuring explicit negative feedback is distinguished from unobserved data. Experiments on the MovieLens 100k and 1M datasets show consistent improvements over state-of-the-art baselines in Precision, NDCG, and MAP while demonstrating robustness for users with limited interaction history. Ablation studies confirm the critical role of LLM-augmented embeddings and the cosine similarity term in capturing nuanced semantic relationships. Our approach effectively mitigates sparsity and cold-start limitations by integrating LLM-derived contextual understanding into graph-based architectures. Future directions include balancing recommendation accuracy with coverage and diversity, and introducing fairness-aware constraints and interpretability features to enhance system performance further.
Fast Graph Neural Network for Image Classification
Aug 20, 2025Abstract:The rapid progress in image classification has been largely driven by the adoption of Graph Convolutional Networks (GCNs), which offer a robust framework for handling complex data structures. This study introduces a novel approach that integrates GCNs with Voronoi diagrams to enhance image classification by leveraging their ability to effectively model relational data. Unlike conventional convolutional neural networks (CNNs), our method represents images as graphs, where pixels or regions function as vertices. These graphs are then refined using corresponding Delaunay triangulations, optimizing their representation. The proposed model achieves significant improvements in both preprocessing efficiency and classification accuracy across various benchmark datasets, surpassing state-of-the-art approaches, particularly in challenging scenarios involving intricate scenes and fine-grained categories. Experimental results, validated through cross-validation, underscore the effectiveness of combining GCNs with Voronoi diagrams for advancing image classification. This research not only presents a novel perspective on image classification but also expands the potential applications of graph-based learning paradigms in computer vision and unstructured data analysis.
Accelerating Image Classification with Graph Convolutional Neural Networks using Voronoi Diagrams
Aug 19, 2025Abstract:Recent advances in image classification have been significantly propelled by the integration of Graph Convolutional Networks (GCNs), offering a novel paradigm for handling complex data structures. This study introduces an innovative framework that employs GCNs in conjunction with Voronoi diagrams to peform image classification, leveraging their exceptional capability to model relational data. Unlike conventional convolutional neural networks, our approach utilizes a graph-based representation of images, where pixels or regions are treated as vertices of a graph, which are then simplified in the form of the corresponding Delaunay triangulations. Our model yields significant improvement in pre-processing time and classification accuracy on several benchmark datasets, surpassing existing state-of-the-art models, especially in scenarios that involve complex scenes and fine-grained categories. The experimental results, validated via cross-validation, underscore the potential of integrating GCNs with Voronoi diagrams in advancing image classification tasks. This research contributes to the field by introducing a novel approach to image classification, while opening new avenues for developing graph-based learning paradigms in other domains of computer vision and non-structured data. In particular, we have proposed a new version of the GCN in this paper, namely normalized Voronoi Graph Convolution Network (NVGCN), which is faster than the regular GCN.
Trustformer: A Trusted Federated Transformer
Jan 20, 2025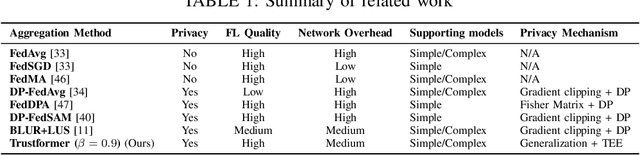
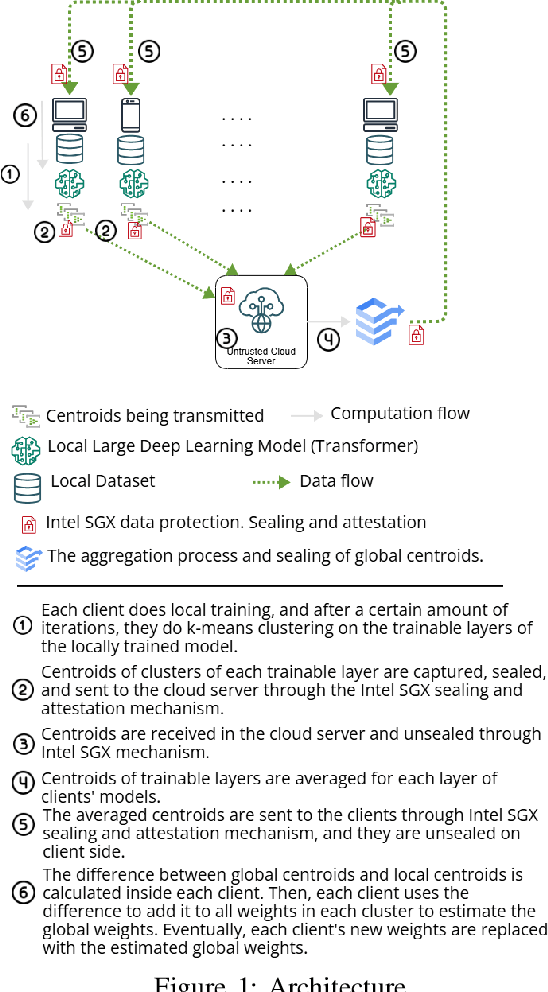
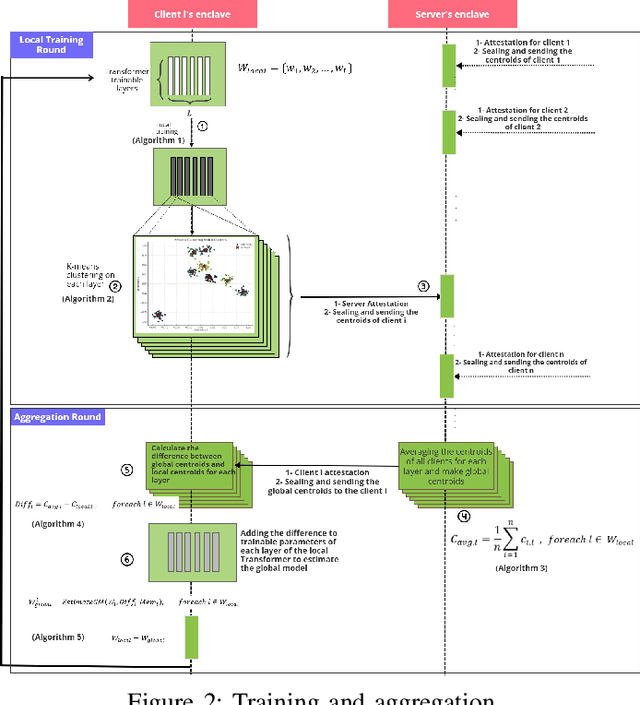

Abstract:Transformers, a cornerstone of deep-learning architectures for sequential data, have achieved state-of-the-art results in tasks like Natural Language Processing (NLP). Models such as BERT and GPT-3 exemplify their success and have driven the rise of large language models (LLMs). However, a critical challenge persists: safeguarding the privacy of data used in LLM training. Privacy-preserving techniques like Federated Learning (FL) offer potential solutions, but practical limitations hinder their effectiveness for Transformer training. Two primary issues are (I) the risk of sensitive information leakage due to aggregation methods like FedAvg or FedSGD, and (II) the high communication overhead caused by the large size of Transformer models. This paper introduces a novel FL method that reduces communication overhead while maintaining competitive utility. Our approach avoids sharing full model weights by simulating a global model locally. We apply k-means clustering to each Transformer layer, compute centroids locally, and transmit only these centroids to the server instead of full weights or gradients. To enhance security, we leverage Intel SGX for secure transmission of centroids. Evaluated on a translation task, our method achieves utility comparable to state-of-the-art baselines while significantly reducing communication costs. This provides a more efficient and privacy-preserving FL solution for Transformer models.
Hypergraph Neural Networks Reveal Spatial Domains from Single-cell Transcriptomics Data
Oct 23, 2024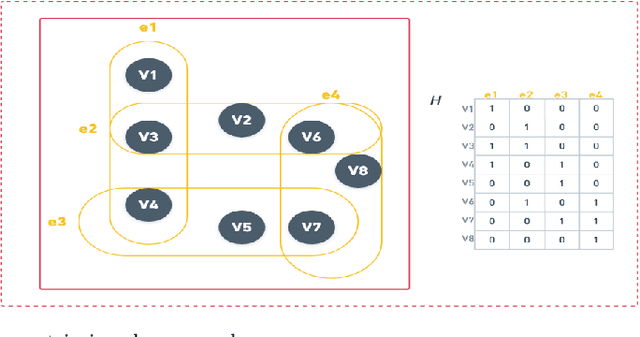
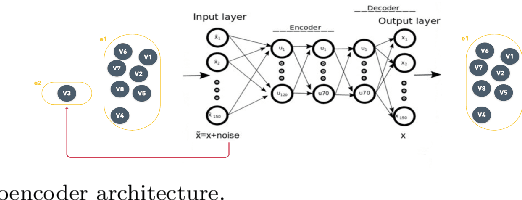
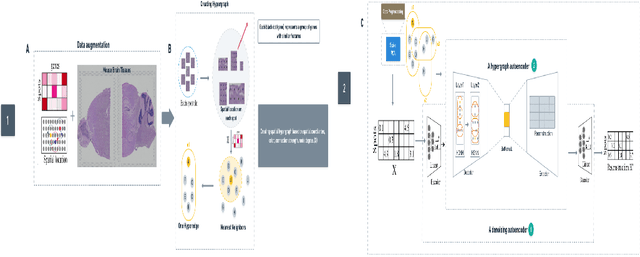
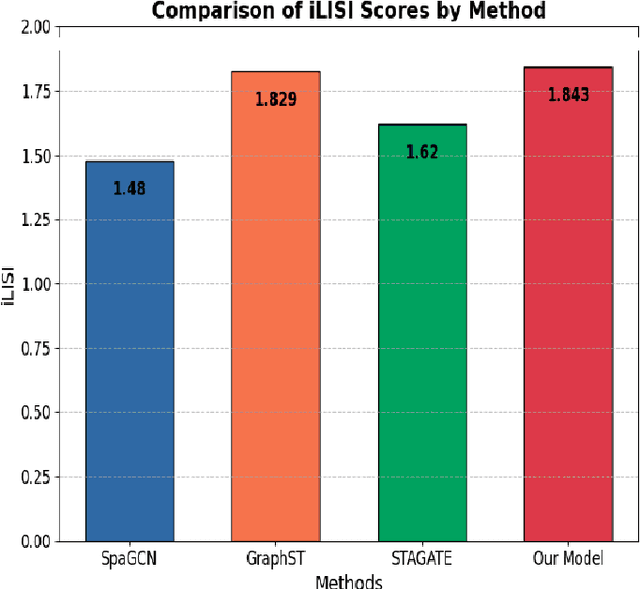
Abstract:The task of spatial clustering of transcriptomics data is of paramount importance. It enables the classification of tissue samples into diverse subpopulations of cells, which, in turn, facilitates the analysis of the biological functions of clusters, tissue reconstruction, and cell-cell interactions. Many approaches leverage gene expressions, spatial locations, and histological images to detect spatial domains; however, Graph Neural Networks (GNNs) as state of the art models suffer from a limitation in the assumption of pairwise connections between nodes. In the case of domain detection in spatial transcriptomics, some cells are found to be not directly related. Still, they are grouped as the same domain, which shows the incapability of GNNs for capturing implicit connections among the cells. While graph edges connect only two nodes, hyperedges connect an arbitrary number of nodes along their edges, which lets Hypergraph Neural Networks (HGNNs) capture and utilize richer and more complex structural information than traditional GNNs. We use autoencoders to address the limitation of not having the actual labels, which are well-suited for unsupervised learning. Our model has demonstrated exceptional performance, achieving the highest iLISI score of 1.843 compared to other methods. This score indicates the greatest diversity of cell types identified by our method. Furthermore, our model outperforms other methods in downstream clustering, achieving the highest ARI values of 0.51 and Leiden score of 0.60.
Hyperedge Modeling in Hypergraph Neural Networks by using Densest Overlapping Subgraphs
Sep 16, 2024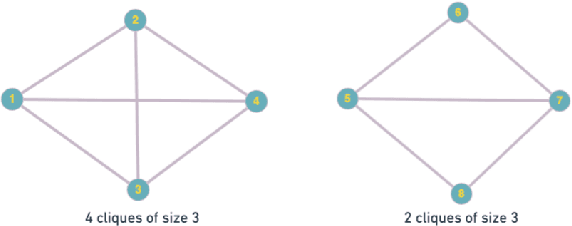
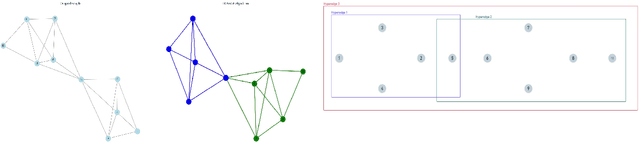
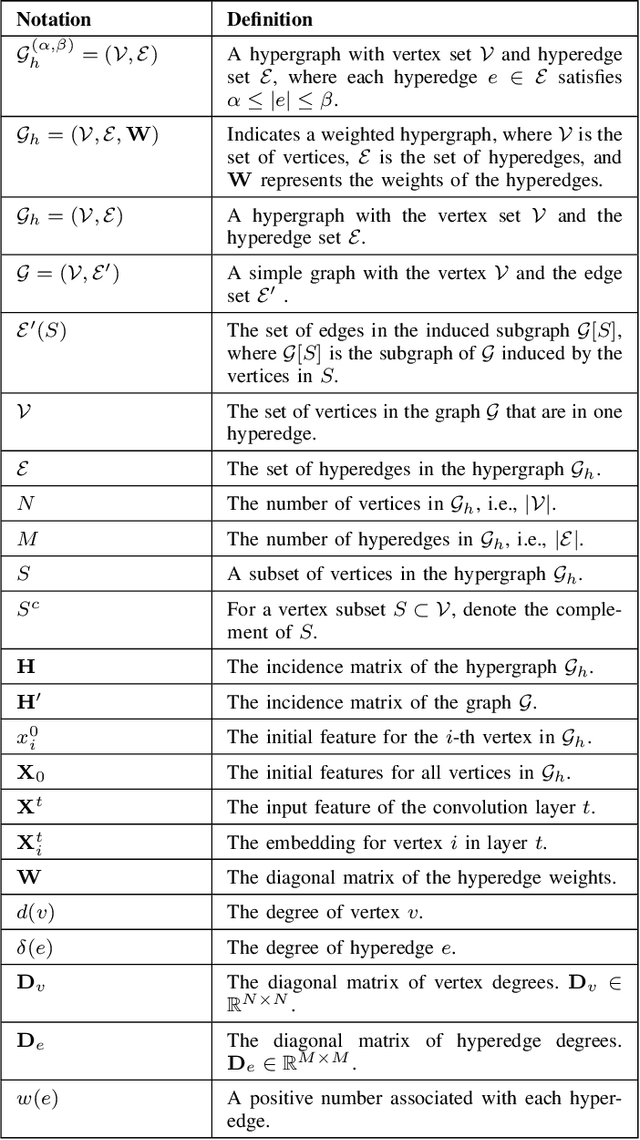
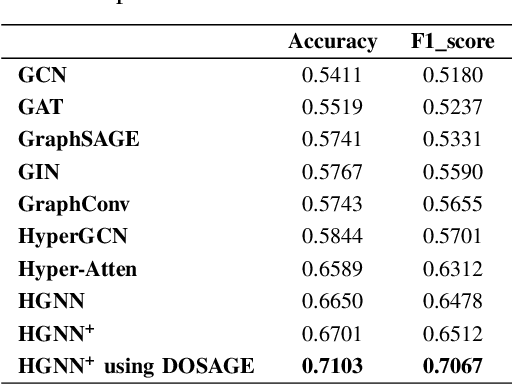
Abstract:Hypergraphs tackle the limitations of traditional graphs by introducing {\em hyperedges}. While graph edges connect only two nodes, hyperedges connect an arbitrary number of nodes along their edges. Also, the underlying message-passing mechanisms in Hypergraph Neural Networks (HGNNs) are in the form of vertex-hyperedge-vertex, which let HGNNs capture and utilize richer and more complex structural information than traditional Graph Neural Networks (GNNs). More recently, the idea of overlapping subgraphs has emerged. These subgraphs can capture more information about subgroups of vertices without limiting one vertex belonging to just one group, allowing vertices to belong to multiple groups or subgraphs. In addition, one of the most important problems in graph clustering is to find densest overlapping subgraphs (DOS). In this paper, we propose a solution to the DOS problem via Agglomerative Greedy Enumeration (DOSAGE) algorithm as a novel approach to enhance the process of generating the densest overlapping subgraphs and, hence, a robust construction of the hypergraphs. Experiments on standard benchmarks show that the DOSAGE algorithm significantly outperforms the HGNNs and six other methods on the node classification task.
Improving Out-of-Distribution Data Handling and Corruption Resistance via Modern Hopfield Networks
Aug 21, 2024Abstract:This study explores the potential of Modern Hopfield Networks (MHN) in improving the ability of computer vision models to handle out-of-distribution data. While current computer vision models can generalize to unseen samples from the same distribution, they are susceptible to minor perturbations such as blurring, which limits their effectiveness in real-world applications. We suggest integrating MHN into the baseline models to enhance their robustness. This integration can be implemented during the test time for any model and combined with any adversarial defense method. Our research shows that the proposed integration consistently improves model performance on the MNIST-C dataset, achieving a state-of-the-art increase of 13.84% in average corruption accuracy, a 57.49% decrease in mean Corruption Error (mCE), and a 60.61% decrease in relative mCE compared to the baseline model. Additionally, we investigate the capability of MHN to converge to the original non-corrupted data. Notably, our method does not require test-time adaptation or augmentation with corruptions, underscoring its practical viability for real-world deployment. (Source code publicly available at: https://github.com/salehsargolzaee/Hopfield-integrated-test)
Lusifer: LLM-based User SImulated Feedback Environment for online Recommender systems
May 22, 2024Abstract:Training reinforcement learning-based recommender systems are often hindered by the lack of dynamic and realistic user interactions. Lusifer, a novel environment leveraging Large Language Models (LLMs), addresses this limitation by generating simulated user feedback. It synthesizes user profiles and interaction histories to simulate responses and behaviors toward recommended items. In addition, user profiles are updated after each rating to reflect evolving user characteristics. Using the MovieLens100K dataset as proof of concept, Lusifer demonstrates accurate emulation of user behavior and preferences. This paper presents Lusifer's operational pipeline, including prompt generation and iterative user profile updates. While validating Lusifer's ability to produce realistic dynamic feedback, future research could utilize this environment to train reinforcement learning systems, offering a scalable and adjustable framework for user simulation in online recommender systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge