Lucas O. Teixeira
Impact of lung segmentation on the diagnosis and explanation of COVID-19 in chest X-ray images
Sep 21, 2020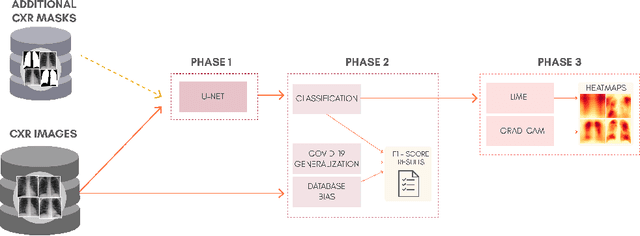
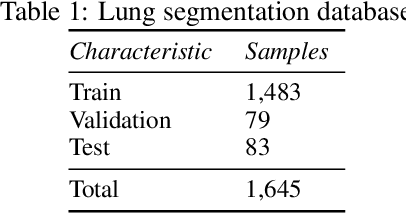
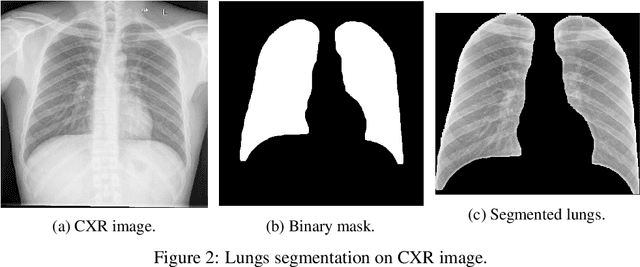
Abstract:The COVID-19 pandemic is undoubtedly one of the biggest public health crises our society has ever faced. This paper's main objectives are to demonstrate the impact of lung segmentation in COVID-19 automatic identification using CXR images and evaluate which contents of the image decisively contribute to the identification. We have performed lung segmentation using a U-Net CNN architecture, and the classification using three well-known CNN architectures: VGG, ResNet, and Inception. To estimate the impact of lung segmentation, we applied some Explainable Artificial Intelligence (XAI), such as LIME and Grad-CAM. To evaluate our approach, we built a database named RYDLS-20-v2, following our previous publication and the COVIDx database guidelines. We evaluated the impact of creating a COVID-19 CXR image database from different sources, called database bias, and the COVID-19 generalization from one database to another, representing our less biased scenario. The experimental results of the segmentation achieved a Jaccard distance of 0.034 and a Dice coefficient of 0.982. In the best and more realistic scenario, we achieved an F1-Score of 0.74 and an area under the ROC curve of 0.9 for COVID-19 identification using segmented CXR images. Further testing and XAI techniques suggest that segmented CXR images represent a much more realistic and less biased performance. More importantly, the experiments conducted show that even after segmentation, there is a strong bias introduced by underlying factors from the data sources, and more efforts regarding the creation of a more significant and comprehensive database still need to be done.
COVID-19 identification in chest X-ray images on flat and hierarchical classification scenarios
May 06, 2020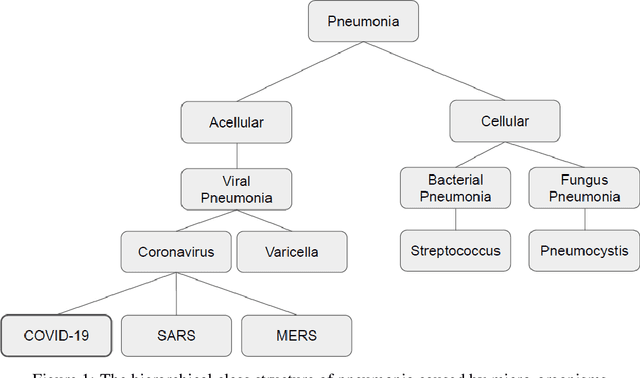

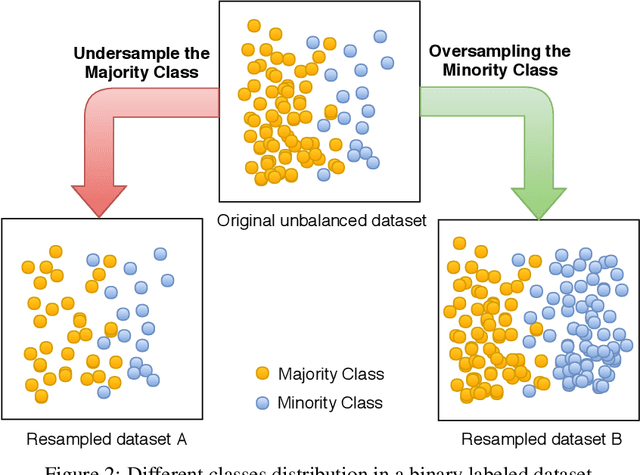
Abstract:The COVID-19 can cause severe pneumonia and is estimated to have a high impact on the healthcare system. The standard image diagnosis tests for pneumonia are chest X-ray (CXR) and computed tomography (CT) scan. CXR are useful in because it is cheaper, faster and more widespread than CT. This study aims to identify pneumonia caused by COVID-19 from other types and also healthy lungs using only CXR images. In order to achieve the objectives, we have proposed a classification schema considering the multi-class and hierarchical perspectives, since pneumonia can be structured as a hierarchy. Given the natural data imbalance in this domain, we also proposed the use of resampling algorithms in order to re-balance the classes distribution. Our classification schema extract features using some well-known texture descriptors and also using a pre-trained CNN model. We also explored early and late fusion techniques in order to leverage the strength of multiple texture descriptors and base classifiers at once. To evaluate the approach, we composed a database, named RYDLS-20, containing CXR images of pneumonia caused by different pathogens as well as CXR images of healthy lungs. The classes distribution follows a real-world scenario in which some pathogens are more common than others. The proposed approach achieved a macro-avg F1-Score of 0.65 using a multi-class approach and a F1-Score of 0.89 for the COVID-19 identification in the hierarchical classification scenario. As far as we know, we achieved the best nominal rate obtained for COVID-19 identification in an unbalanced environment with more than three classes. We must also highlight the novel proposed hierarchical classification approach for this task, which considers the types of pneumonia caused by the different pathogens and lead us to the best COVID-19 recognition rate obtained here.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge