Luca Pasquali
Deep Generative Markov State Models
May 19, 2018
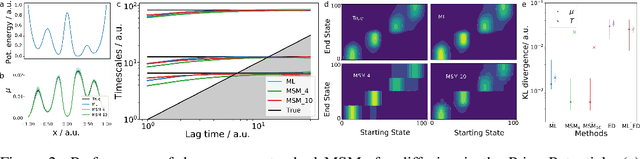
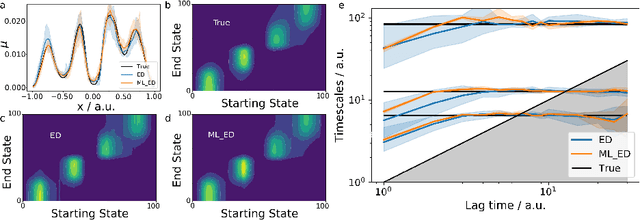
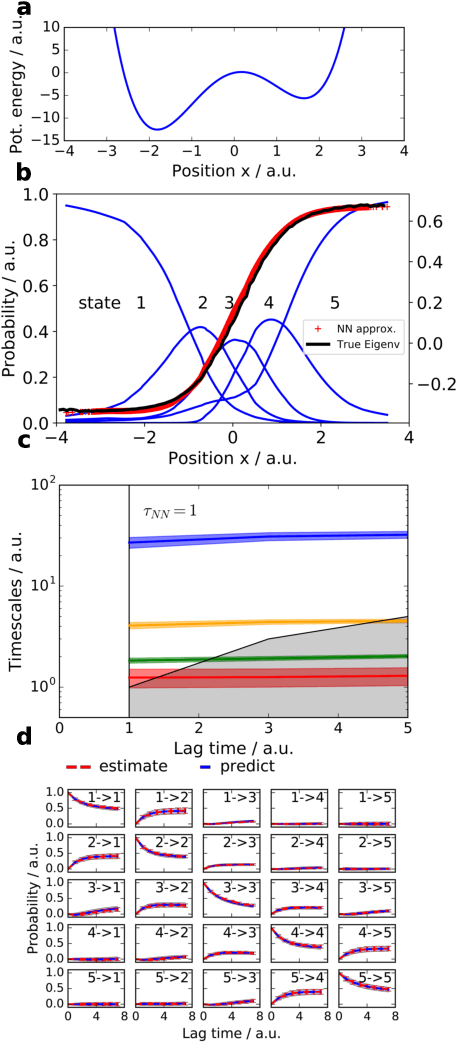
Abstract:We propose a deep generative Markov State Model (DeepGenMSM) learning framework for inference of metastable dynamical systems and prediction of trajectories. After unsupervised training on time series data, the model contains (i) a probabilistic encoder that maps from high-dimensional configuration space to a small-sized vector indicating the membership to metastable (long-lived) states, (ii) a Markov chain that governs the transitions between metastable states and facilitates analysis of the long-time dynamics, and (iii) a generative part that samples the conditional distribution of configurations in the next time step. The model can be operated in a recursive fashion to generate trajectories to predict the system evolution from a defined starting state and propose new configurations. The DeepGenMSM is demonstrated to provide accurate estimates of the long-time kinetics and generate valid distributions for molecular dynamics (MD) benchmark systems. Remarkably, we show that DeepGenMSMs are able to make long time-steps in molecular configuration space and generate physically realistic structures in regions that were not seen in training data.
VAMPnets: Deep learning of molecular kinetics
Dec 20, 2017


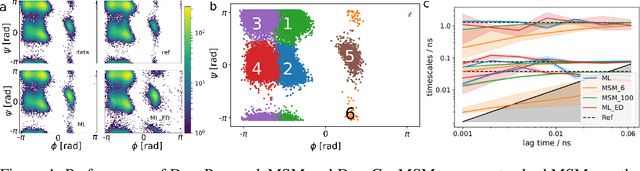
Abstract:There is an increasing demand for computing the relevant structures, equilibria and long-timescale kinetics of biomolecular processes, such as protein-drug binding, from high-throughput molecular dynamics simulations. Current methods employ transformation of simulated coordinates into structural features, dimension reduction, clustering the dimension-reduced data, and estimation of a Markov state model or related model of the interconversion rates between molecular structures. This handcrafted approach demands a substantial amount of modeling expertise, as poor decisions at any step will lead to large modeling errors. Here we employ the variational approach for Markov processes (VAMP) to develop a deep learning framework for molecular kinetics using neural networks, dubbed VAMPnets. A VAMPnet encodes the entire mapping from molecular coordinates to Markov states, thus combining the whole data processing pipeline in a single end-to-end framework. Our method performs equally or better than state-of-the art Markov modeling methods and provides easily interpretable few-state kinetic models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge