Louis D. van Harten
Task-Based Adaptive Transmit Beamforming for Efficient Ultrasound Quantification
Jan 28, 2026Abstract:Wireless and wearable ultrasound devices promise to enable continuous ultrasound monitoring, but power consumption and data throughput remain critical challenges. Reducing the number of transmit events per second directly impacts both. We propose a task-based adaptive transmit beamforming method, formulated as a Bayesian active perception problem, that adaptively chooses where to scan in order to gain information about downstream quantitative measurements, avoiding redundant transmit events. Our proposed Task-Based Information Gain (TBIG) strategy applies to any differentiable downstream task function. When applied to recovering ventricular dimensions from echocardiograms, TBIG recovers accurate results using fewer than 2% of scan lines typically used, showing potential for large reductions in the power usage and data rates necessary for monitoring. Code is available at https://github.com/tue-bmd/task-based-ulsa.
Robust deformable image registration using cycle-consistent implicit representations
Oct 03, 2023Abstract:Recent works in medical image registration have proposed the use of Implicit Neural Representations, demonstrating performance that rivals state-of-the-art learning-based methods. However, these implicit representations need to be optimized for each new image pair, which is a stochastic process that may fail to converge to a global minimum. To improve robustness, we propose a deformable registration method using pairs of cycle-consistent Implicit Neural Representations: each implicit representation is linked to a second implicit representation that estimates the opposite transformation, causing each network to act as a regularizer for its paired opposite. During inference, we generate multiple deformation estimates by numerically inverting the paired backward transformation and evaluating the consensus of the optimized pair. This consensus improves registration accuracy over using a single representation and results in a robust uncertainty metric that can be used for automatic quality control. We evaluate our method with a 4D lung CT dataset. The proposed cycle-consistent optimization method reduces the optimization failure rate from 2.4% to 0.0% compared to the current state-of-the-art. The proposed inference method improves landmark accuracy by 4.5% and the proposed uncertainty metric detects all instances where the registration method fails to converge to a correct solution. We verify the generalizability of these results to other data using a centerline propagation task in abdominal 4D MRI, where our method achieves a 46% improvement in propagation consistency compared with single-INR registration and demonstrates a strong correlation between the proposed uncertainty metric and registration accuracy.
Automatic Online Quality Control of Synthetic CTs
Nov 12, 2019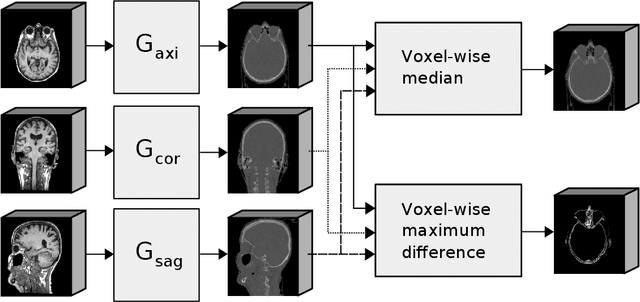
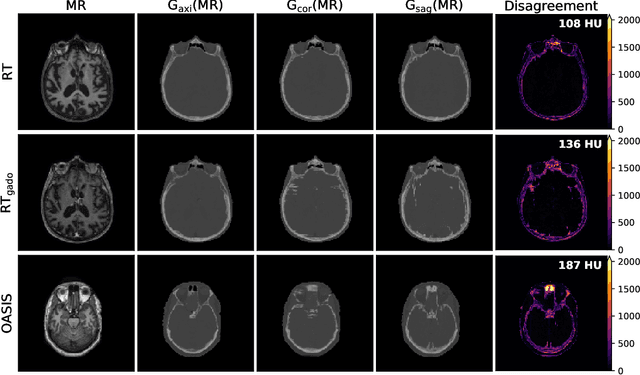
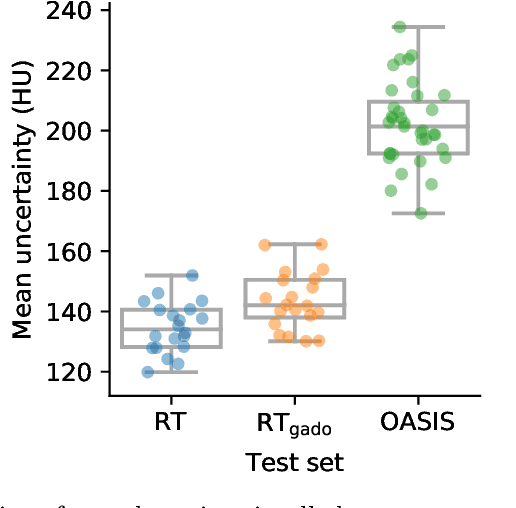
Abstract:Accurate MR-to-CT synthesis is a requirement for MR-only workflows in radiotherapy (RT) treatment planning. In recent years, deep learning-based approaches have shown impressive results in this field. However, to prevent downstream errors in RT treatment planning, it is important that deep learning models are only applied to data for which they are trained and that generated synthetic CT (sCT) images do not contain severe errors. For this, a mechanism for online quality control should be in place. In this work, we use an ensemble of sCT generators and assess their disagreement as a measure of uncertainty of the results. We show that this uncertainty measure can be used for two kinds of online quality control. First, to detect input images that are outside the expected distribution of MR images. Second, to identify sCT images that were generated from suitable MR images but potentially contain errors. Such automatic online quality control for sCT generation is likely to become an integral part of MR-only RT workflows.
Exploiting Clinically Available Delineations for CNN-based Segmentation in Radiotherapy Treatment Planning
Nov 12, 2019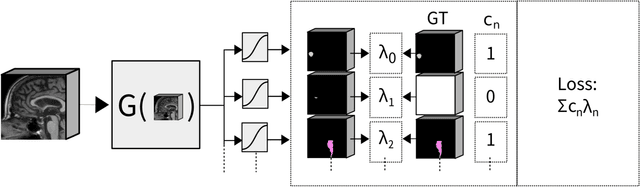
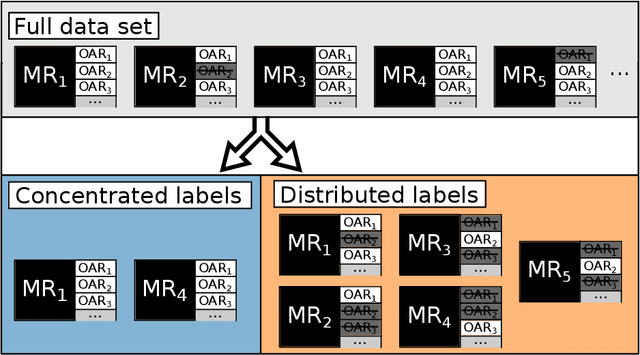
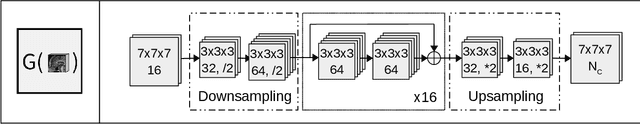
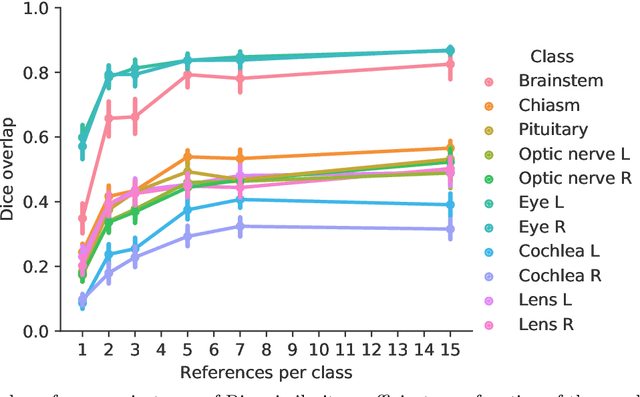
Abstract:Convolutional neural networks (CNNs) have been widely and successfully used for medical image segmentation. However, CNNs are typically considered to require large numbers of dedicated expert-segmented training volumes, which may be limiting in practice. This work investigates whether clinically obtained segmentations which are readily available in picture archiving and communication systems (PACS) could provide a possible source of data to train a CNN for segmentation of organs-at-risk (OARs) in radiotherapy treatment planning. In such data, delineations of structures deemed irrelevant to the target clinical use may be lacking. To overcome this issue, we use multi-label instead of multi-class segmentation. We empirically assess how many clinical delineations would be sufficient to train a CNN for the segmentation of OARs and find that increasing the training set size beyond a limited number of images leads to sharply diminishing returns. Moreover, we find that by using multi-label segmentation, missing structures in the reference standard do not have a negative effect on overall segmentation accuracy. These results indicate that segmentations obtained in a clinical workflow can be used to train an accurate OAR segmentation model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge