Lorenzo Simone
Interpretable Early Detection of Parkinson's Disease through Speech Analysis
Apr 24, 2025
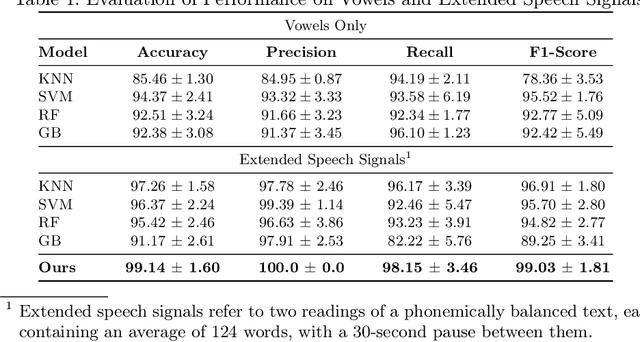

Abstract:Parkinson's disease is a progressive neurodegenerative disorder affecting motor and non-motor functions, with speech impairments among its earliest symptoms. Speech impairments offer a valuable diagnostic opportunity, with machine learning advances providing promising tools for timely detection. In this research, we propose a deep learning approach for early Parkinson's disease detection from speech recordings, which also highlights the vocal segments driving predictions to enhance interpretability. This approach seeks to associate predictive speech patterns with articulatory features, providing a basis for interpreting underlying neuromuscular impairments. We evaluated our approach using the Italian Parkinson's Voice and Speech Database, containing 831 audio recordings from 65 participants, including both healthy individuals and patients. Our approach showed competitive classification performance compared to state-of-the-art methods, while providing enhanced interpretability by identifying key speech features influencing predictions.
Towards Efficient Molecular Property Optimization with Graph Energy Based Models
Feb 17, 2025



Abstract:Optimizing chemical properties is a challenging task due to the vastness and complexity of chemical space. Here, we present a generative energy-based architecture for implicit chemical property optimization, designed to efficiently generate molecules that satisfy target properties without explicit conditional generation. We use Graph Energy Based Models and a training approach that does not require property labels. We validated our approach on well-established chemical benchmarks, showing superior results to state-of-the-art methods and demonstrating robustness and efficiency towards de novo drug design.
On the effectiveness of smartphone IMU sensors and Deep Learning in the detection of cardiorespiratory conditions
Aug 27, 2024Abstract:This research introduces an innovative method for the early screening of cardiorespiratory diseases based on an acquisition protocol, which leverages commodity smartphone's Inertial Measurement Units (IMUs) and deep learning techniques. We collected, in a clinical setting, a dataset featuring recordings of breathing kinematics obtained by accelerometer and gyroscope readings from five distinct body regions. We propose an end-to-end deep learning pipeline for early cardiorespiratory disease screening, incorporating a preprocessing step segmenting the data into individual breathing cycles, and a recurrent bidirectional module capturing features from diverse body regions. We employed Leave-one-out-cross-validation with Bayesian optimization for hyperparameter tuning and model selection. The experimental results consistently demonstrated the superior performance of a bidirectional Long-Short Term Memory (Bi-LSTM) as a feature encoder architecture, yielding an average sensitivity of $0.81 \pm 0.02$, specificity of $0.82 \pm 0.05$, F1 score of $0.81 \pm 0.02$, and accuracy of $80.2\% \pm 3.9$ across diverse seed variations. We also assessed generalization capabilities on a skewed distribution, comprising exclusively healthy patients not used in training, revealing a true negative rate of $74.8 \% \pm 4.5$. The sustained accuracy of predictions over time during breathing cycles within a single patient underscores the efficacy of the preprocessing strategy, highlighting the model's ability to discern significant patterns throughout distinct phases of the respiratory cycle. This investigation underscores the potential usefulness of widely available smartphones as devices for timely cardiorespiratory disease screening in the general population, in at-home settings, offering crucial assistance to public health efforts (especially during a pandemic outbreaks, such as the recent COVID-19).
ECGAN: Self-supervised generative adversarial network for electrocardiography
Jan 23, 2023Abstract:High-quality synthetic data can support the development of effective predictive models for biomedical tasks, especially in rare diseases or when subject to compelling privacy constraints. These limitations, for instance, negatively impact open access to electrocardiography datasets about arrhythmias. This work introduces a self-supervised approach to the generation of synthetic electrocardiography time series which is shown to promote morphological plausibility. Our model (ECGAN) allows conditioning the generative process for specific rhythm abnormalities, enhancing synchronization and diversity across samples with respect to literature models. A dedicated sample quality assessment framework is also defined, leveraging arrhythmia classifiers. The empirical results highlight a substantial improvement against state-of-the-art generative models for sequences and audio synthesis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge