Litao Qiao
Towards Explainable Automated Neuroanatomy
Apr 08, 2024Abstract:We present a novel method for quantifying the microscopic structure of brain tissue. It is based on the automated recognition of interpretable features obtained by analyzing the shapes of cells. This contrasts with prevailing methods of brain anatomical analysis in two ways. First, contemporary methods use gray-scale values derived from smoothed version of the anatomical images, which dissipated valuable information from the texture of the images. Second, contemporary analysis uses the output of black-box Convolutional Neural Networks, while our system makes decisions based on interpretable features obtained by analyzing the shapes of individual cells. An important benefit of this open-box approach is that the anatomist can understand and correct the decisions made by the computer. Our proposed system can accurately localize and identify existing brain structures. This can be used to align and coregistar brains and will facilitate connectomic studies for reverse engineering of brain circuitry.
Learning Accurate and Interpretable Decision Rule Sets from Neural Networks
Mar 12, 2021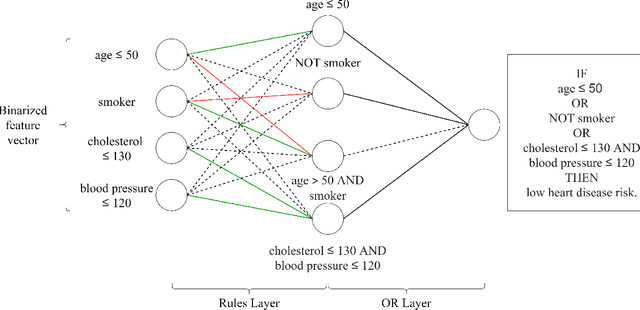
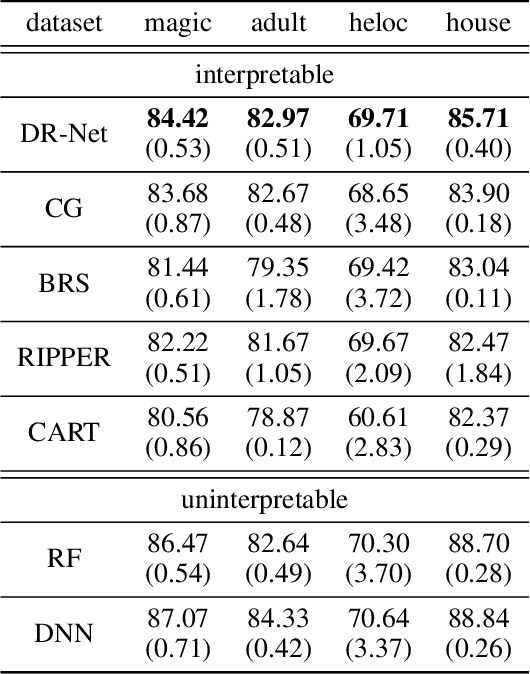
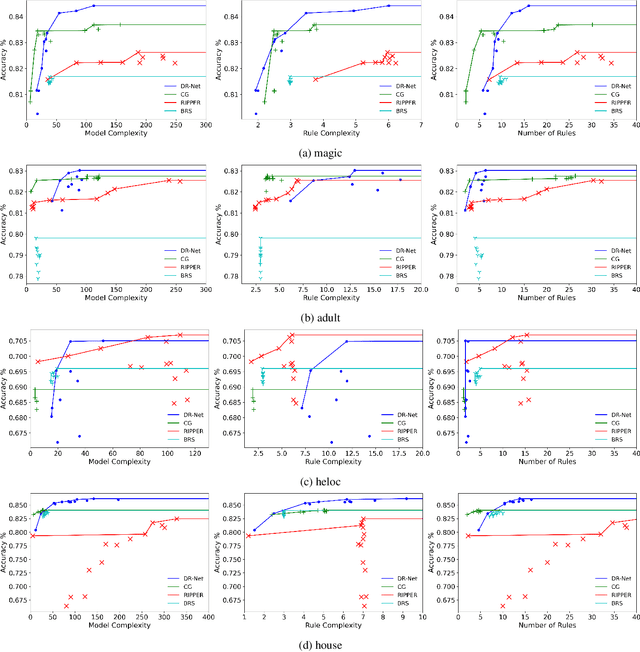

Abstract:This paper proposes a new paradigm for learning a set of independent logical rules in disjunctive normal form as an interpretable model for classification. We consider the problem of learning an interpretable decision rule set as training a neural network in a specific, yet very simple two-layer architecture. Each neuron in the first layer directly maps to an interpretable if-then rule after training, and the output neuron in the second layer directly maps to a disjunction of the first-layer rules to form the decision rule set. Our representation of neurons in this first rules layer enables us to encode both the positive and the negative association of features in a decision rule. State-of-the-art neural net training approaches can be leveraged for learning highly accurate classification models. Moreover, we propose a sparsity-based regularization approach to balance between classification accuracy and the simplicity of the derived rules. Our experimental results show that our method can generate more accurate decision rule sets than other state-of-the-art rule-learning algorithms with better accuracy-simplicity trade-offs. Further, when compared with uninterpretable black-box machine learning approaches such as random forests and full-precision deep neural networks, our approach can easily find interpretable decision rule sets that have comparable predictive performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge