Lior Golgher
Reconstructing the Hemodynamic Response Function via a Bimodal Transformer
Jun 28, 2023



Abstract:The relationship between blood flow and neuronal activity is widely recognized, with blood flow frequently serving as a surrogate for neuronal activity in fMRI studies. At the microscopic level, neuronal activity has been shown to influence blood flow in nearby blood vessels. This study introduces the first predictive model that addresses this issue directly at the explicit neuronal population level. Using in vivo recordings in awake mice, we employ a novel spatiotemporal bimodal transformer architecture to infer current blood flow based on both historical blood flow and ongoing spontaneous neuronal activity. Our findings indicate that incorporating neuronal activity significantly enhances the model's ability to predict blood flow values. Through analysis of the model's behavior, we propose hypotheses regarding the largely unexplored nature of the hemodynamic response to neuronal activity.
Microvascular Dynamics from 4D Microscopy Using Temporal Segmentation
Jan 14, 2020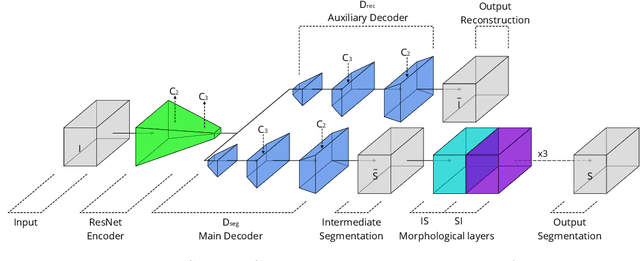


Abstract:Recently developed methods for rapid continuous volumetric two-photon microscopy facilitate the observation of neuronal activity in hundreds of individual neurons and changes in blood flow in adjacent blood vessels across a large volume of living brain at unprecedented spatio-temporal resolution. However, the high imaging rate necessitates fully automated image analysis, whereas tissue turbidity and photo-toxicity limitations lead to extremely sparse and noisy imagery. In this work, we extend a recently proposed deep learning volumetric blood vessel segmentation network, such that it supports temporal analysis. With this technology, we are able to track changes in cerebral blood volume over time and identify spontaneous arterial dilations that propagate towards the pial surface. This new capability is a promising step towards characterizing the hemodynamic response function upon which functional magnetic resonance imaging (fMRI) is based.
Unsupervised Microvascular Image Segmentation Using an Active Contours Mimicking Neural Network
Aug 16, 2019
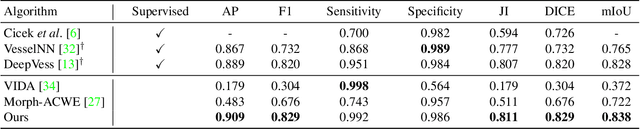
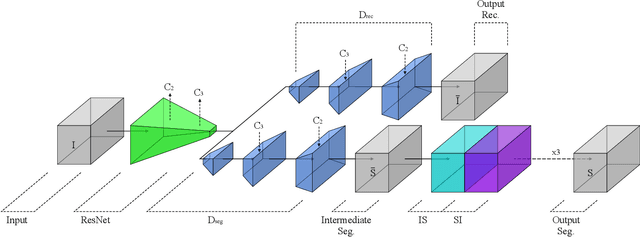
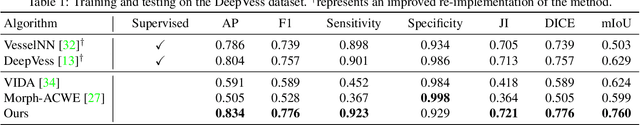
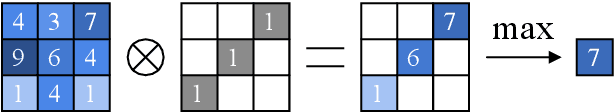
Abstract:The task of blood vessel segmentation in microscopy images is crucial for many diagnostic and research applications. However, vessels can look vastly different, depending on the transient imaging conditions, and collecting data for supervised training is laborious. We present a novel deep learning method for unsupervised segmentation of blood vessels. The method is inspired by the field of active contours and we introduce a new loss term, which is based on the morphological Active Contours Without Edges (ACWE) optimization method. The role of the morphological operators is played by novel pooling layers that are incorporated to the network's architecture. We demonstrate the challenges that are faced by previous supervised learning solutions, when the imaging conditions shift. Our unsupervised method is able to outperform such previous methods in both the labeled dataset, and when applied to similar but different datasets. Our code, as well as efficient PyTorch reimplementations of the baseline methods VesselNN and DeepVess is available on GitHub - https://github.com/shirgur/UMIS.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge