Lingli Yang
On the Confidence Intervals in Bioequivalence Studies
Jun 11, 2023Abstract:A bioequivalence study is a type of clinical trial designed to compare the biological equivalence of two different formulations of a drug. Such studies are typically conducted in controlled clinical settings with human subjects, who are randomly assigned to receive two formulations. The two formulations are then compared with respect to their pharmacokinetic profiles, which encompass the absorption, distribution, metabolism, and elimination of the drug. Under the guidance from Food and Drug Administration (FDA), for a size-$\alpha$ bioequivalence test, the standard approach is to construct a $100(1-2\alpha)\%$ confidence interval and verify if the confidence interval falls with the critical region. In this work, we clarify that $100(1-2\alpha)\%$ confidence interval approach for bioequivalence testing yields a size-$\alpha$ test only when the two one-sided tests in TOST are ``equal-tailed''. Furthermore, a $100(1-\alpha)\%$ confidence interval approach is also discussed in the bioequivalence study.
Deep Feature Screening: Feature Selection for Ultra High-Dimensional Data via Deep Neural Networks
Apr 04, 2022
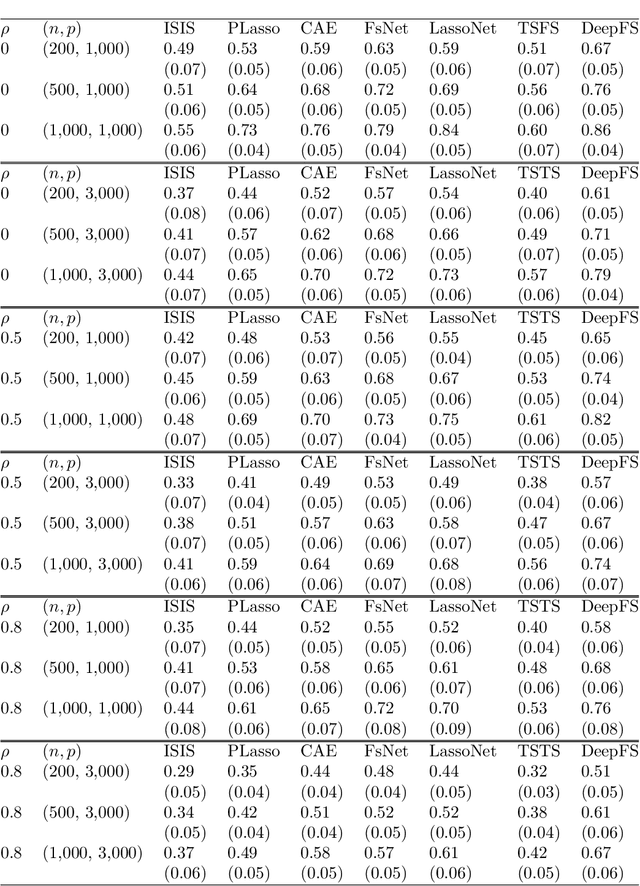
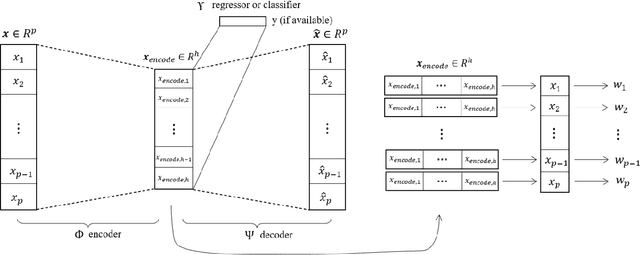
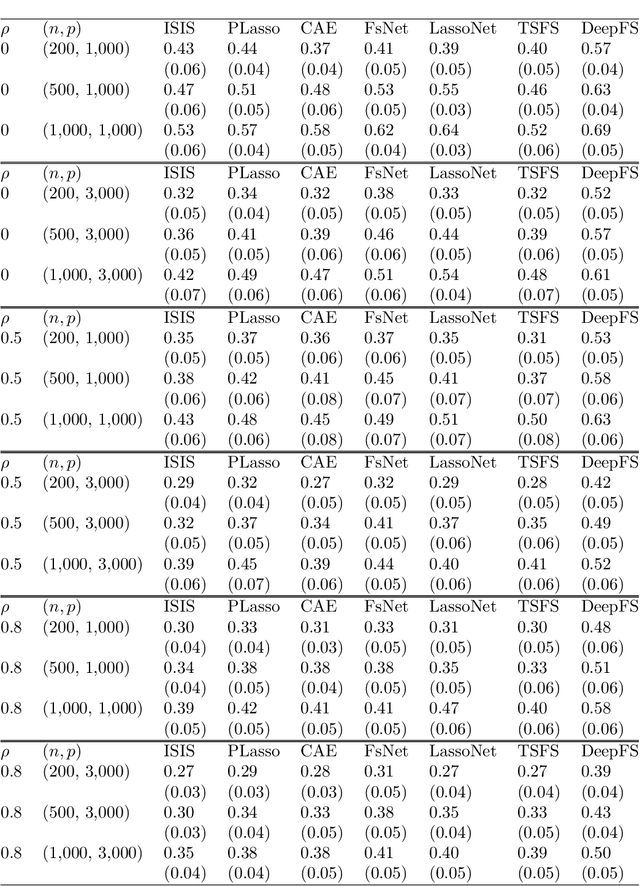
Abstract:The applications of traditional statistical feature selection methods to high-dimension, low sample-size data often struggle and encounter challenging problems, such as overfitting, curse of dimensionality, computational infeasibility, and strong model assumption. In this paper, we propose a novel two-step nonparametric approach called Deep Feature Screening (DeepFS) that can overcome these problems and identify significant features with high precision for ultra high-dimensional, low-sample-size data. This approach first extracts a low-dimensional representation of input data and then applies feature screening based on multivariate rank distance correlation recently developed by Deb and Sen (2021). This approach combines the strengths of both deep neural networks and feature screening, and thereby has the following appealing features in addition to its ability of handling ultra high-dimensional data with small number of samples: (1) it is model free and distribution free; (2) it can be used for both supervised and unsupervised feature selection; and (3) it is capable of recovering the original input data. The superiority of DeepFS is demonstrated via extensive simulation studies and real data analyses.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge