Lena Schmidt
Horizon Scans can be accelerated using novel information retrieval and artificial intelligence tools
Apr 02, 2025Abstract:Introduction: Horizon scanning in healthcare assesses early signals of innovation, crucial for timely adoption. Current horizon scanning faces challenges in efficient information retrieval and analysis, especially from unstructured sources like news, presenting a need for innovative tools. Methodology: The study introduces SCANAR and AIDOC, open-source Python-based tools designed to improve horizon scanning. SCANAR automates the retrieval and processing of news articles, offering functionalities such as de-duplication and unsupervised relevancy ranking. AIDOC aids filtration by leveraging AI to reorder textual data based on relevancy, employing neural networks for semantic similarity, and subsequently prioritizing likely relevant entries for human review. Results: Twelve internal datasets from horizon scans and four external benchmarking datasets were used. SCANAR improved retrieval efficiency by automating processes previously dependent on manual labour. AIDOC displayed work-saving potential, achieving around 62% reduction in manual review efforts at 95% recall. Comparative analysis with benchmarking data showed AIDOC's performance was similar to existing systematic review automation tools, though performance varied depending on dataset characteristics. A smaller case-study on our news datasets shows the potential of ensembling large language models within the active-learning process for faster detection of relevant articles across news datasets. Conclusion: The validation indicates that SCANAR and AIDOC show potential to enhance horizon scanning efficiency by streamlining data retrieval and prioritisation. These tools may alleviate methodological limitations and allow broader, swifter horizon scans. Further studies are suggested to optimize these models and to design new workflows and validation processes that integrate large language models.
Exploring the use of a Large Language Model for data extraction in systematic reviews: a rapid feasibility study
May 23, 2024Abstract:This paper describes a rapid feasibility study of using GPT-4, a large language model (LLM), to (semi)automate data extraction in systematic reviews. Despite the recent surge of interest in LLMs there is still a lack of understanding of how to design LLM-based automation tools and how to robustly evaluate their performance. During the 2023 Evidence Synthesis Hackathon we conducted two feasibility studies. Firstly, to automatically extract study characteristics from human clinical, animal, and social science domain studies. We used two studies from each category for prompt-development; and ten for evaluation. Secondly, we used the LLM to predict Participants, Interventions, Controls and Outcomes (PICOs) labelled within 100 abstracts in the EBM-NLP dataset. Overall, results indicated an accuracy of around 80%, with some variability between domains (82% for human clinical, 80% for animal, and 72% for studies of human social sciences). Causal inference methods and study design were the data extraction items with the most errors. In the PICO study, participants and intervention/control showed high accuracy (>80%), outcomes were more challenging. Evaluation was done manually; scoring methods such as BLEU and ROUGE showed limited value. We observed variability in the LLMs predictions and changes in response quality. This paper presents a template for future evaluations of LLMs in the context of data extraction for systematic review automation. Our results show that there might be value in using LLMs, for example as second or third reviewers. However, caution is advised when integrating models such as GPT-4 into tools. Further research on stability and reliability in practical settings is warranted for each type of data that is processed by the LLM.
* Conference proceedings, peer-reviewed and presented at the 3rd Workshop on Augmented Intelligence for Technology-Assisted Reviews Systems, Glasgow, 2024
Data Mining in Clinical Trial Text: Transformers for Classification and Question Answering Tasks
Jan 30, 2020


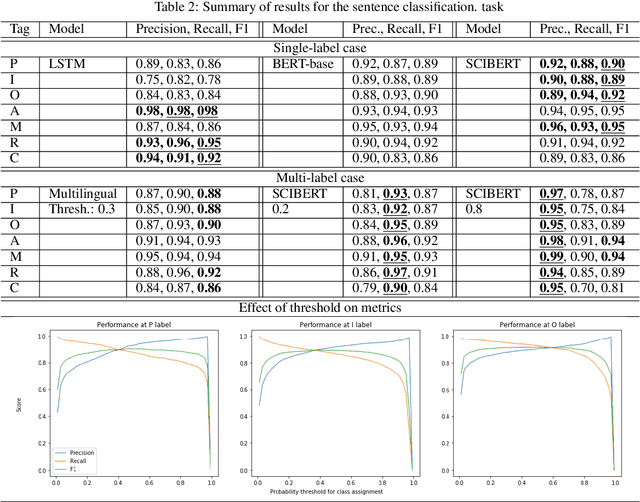
Abstract:This research on data extraction methods applies recent advances in natural language processing to evidence synthesis based on medical texts. Texts of interest include abstracts of clinical trials in English and in multilingual contexts. The main focus is on information characterized via the Population, Intervention, Comparator, and Outcome (PICO) framework, but data extraction is not limited to these fields. Recent neural network architectures based on transformers show capacities for transfer learning and increased performance on downstream natural language processing tasks such as universal reading comprehension, brought forward by this architecture's use of contextualized word embeddings and self-attention mechanisms. This paper contributes to solving problems related to ambiguity in PICO sentence prediction tasks, as well as highlighting how annotations for training named entity recognition systems are used to train a high-performing, but nevertheless flexible architecture for question answering in systematic review automation. Additionally, it demonstrates how the problem of insufficient amounts of training annotations for PICO entity extraction is tackled by augmentation. All models in this paper were created with the aim to support systematic review (semi)automation. They achieve high F1 scores, and demonstrate the feasibility of applying transformer-based classification methods to support data mining in the biomedical literature.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge