Lei Jimmy Ba
Classifying and Segmenting Microscopy Images Using Convolutional Multiple Instance Learning
Nov 17, 2015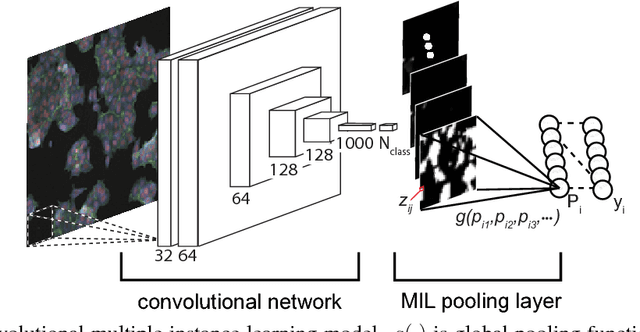
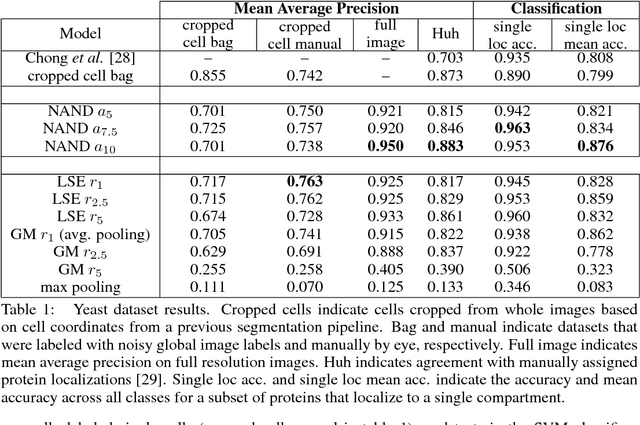
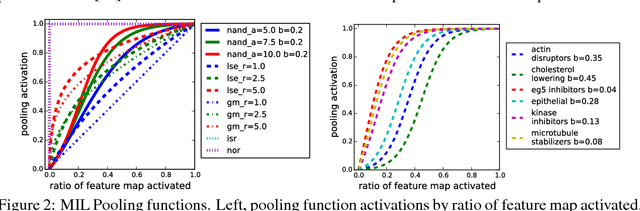
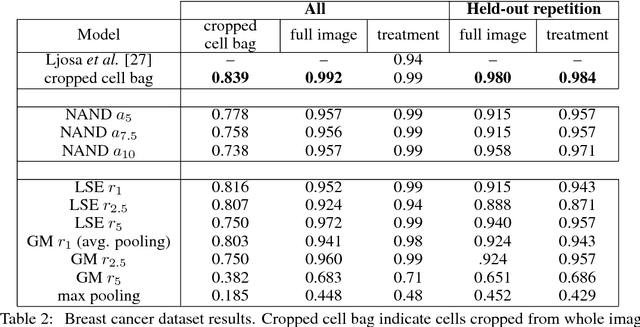
Abstract:Convolutional neural networks (CNN) have achieved state of the art performance on both classification and segmentation tasks. Applying CNNs to microscopy images is challenging due to the lack of datasets labeled at the single cell level. We extend the application of CNNs to microscopy image classification and segmentation using multiple instance learning (MIL). We present the adaptive Noisy-AND MIL pooling function, a new MIL operator that is robust to outliers. Combining CNNs with MIL enables training CNNs using full resolution microscopy images with global labels. We base our approach on the similarity between the aggregation function used in MIL and pooling layers used in CNNs. We show that training MIL CNNs end-to-end outperforms several previous methods on both mammalian and yeast microscopy images without requiring any segmentation steps.
Do Deep Nets Really Need to be Deep?
Oct 11, 2014
Abstract:Currently, deep neural networks are the state of the art on problems such as speech recognition and computer vision. In this extended abstract, we show that shallow feed-forward networks can learn the complex functions previously learned by deep nets and achieve accuracies previously only achievable with deep models. Moreover, in some cases the shallow neural nets can learn these deep functions using a total number of parameters similar to the original deep model. We evaluate our method on the TIMIT phoneme recognition task and are able to train shallow fully-connected nets that perform similarly to complex, well-engineered, deep convolutional architectures. Our success in training shallow neural nets to mimic deeper models suggests that there probably exist better algorithms for training shallow feed-forward nets than those currently available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge