Classifying and Segmenting Microscopy Images Using Convolutional Multiple Instance Learning
Paper and Code
Nov 17, 2015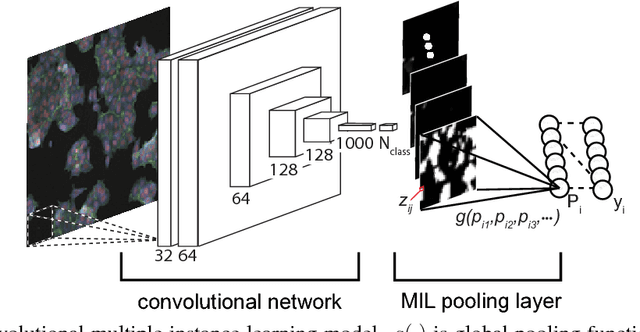
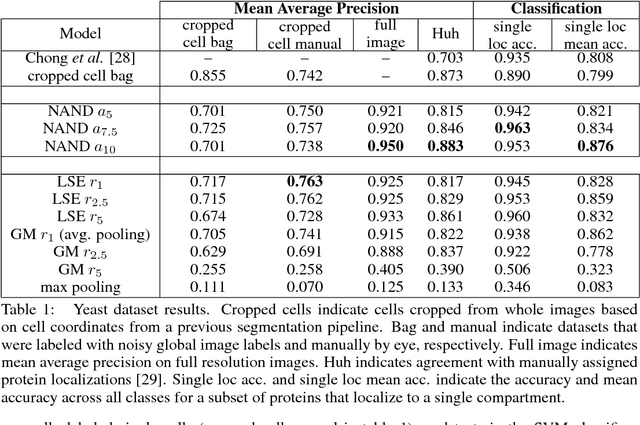
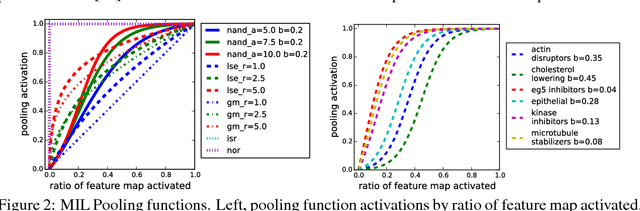
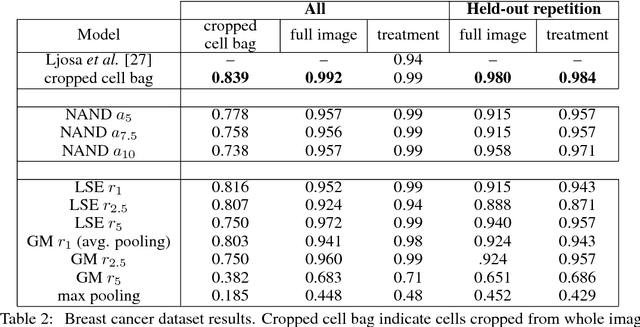
Convolutional neural networks (CNN) have achieved state of the art performance on both classification and segmentation tasks. Applying CNNs to microscopy images is challenging due to the lack of datasets labeled at the single cell level. We extend the application of CNNs to microscopy image classification and segmentation using multiple instance learning (MIL). We present the adaptive Noisy-AND MIL pooling function, a new MIL operator that is robust to outliers. Combining CNNs with MIL enables training CNNs using full resolution microscopy images with global labels. We base our approach on the similarity between the aggregation function used in MIL and pooling layers used in CNNs. We show that training MIL CNNs end-to-end outperforms several previous methods on both mammalian and yeast microscopy images without requiring any segmentation steps.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge