Lea Goetz
Individual Fairness Through Reweighting and Tuning
May 07, 2024

Abstract:Inherent bias within society can be amplified and perpetuated by artificial intelligence (AI) systems. To address this issue, a wide range of solutions have been proposed to identify and mitigate bias and enforce fairness for individuals and groups. Recently, Graph Laplacian Regularizer (GLR), a regularization technique from the semi-supervised learning literature has been used as a substitute for the common Lipschitz condition to enhance individual fairness. Notable prior work has shown that enforcing individual fairness through a GLR can improve the transfer learning accuracy of AI models under covariate shifts. However, the prior work defines a GLR on the source and target data combined, implicitly assuming that the target data are available at train time, which might not hold in practice. In this work, we investigated whether defining a GLR independently on the train and target data could maintain similar accuracy. Furthermore, we introduced the Normalized Fairness Gain score (NFG) to measure individual fairness by measuring the amount of gained fairness when a GLR is used versus not. We evaluated the new and original methods under NFG, the Prediction Consistency (PC), and traditional classification metrics on the German Credit Approval dataset. The results showed that the two models achieved similar statistical mean performances over five-fold cross-validation. Furthermore, the proposed metric showed that PC scores can be misleading as the scores can be high and statistically similar to fairness-enhanced models while NFG scores are small. This work therefore provides new insights into when a GLR effectively enhances individual fairness and the pitfalls of PC.
RAmBLA: A Framework for Evaluating the Reliability of LLMs as Assistants in the Biomedical Domain
Mar 21, 2024



Abstract:Large Language Models (LLMs) increasingly support applications in a wide range of domains, some with potential high societal impact such as biomedicine, yet their reliability in realistic use cases is under-researched. In this work we introduce the Reliability AssesMent for Biomedical LLM Assistants (RAmBLA) framework and evaluate whether four state-of-the-art foundation LLMs can serve as reliable assistants in the biomedical domain. We identify prompt robustness, high recall, and a lack of hallucinations as necessary criteria for this use case. We design shortform tasks and tasks requiring LLM freeform responses mimicking real-world user interactions. We evaluate LLM performance using semantic similarity with a ground truth response, through an evaluator LLM.
On generalisability of segment anything model for nuclear instance segmentation in histology images
Jan 25, 2024Abstract:Pre-trained on a large and diverse dataset, the segment anything model (SAM) is the first promptable foundation model in computer vision aiming at object segmentation tasks. In this work, we evaluate SAM for the task of nuclear instance segmentation performance with zero-shot learning and finetuning. We compare SAM with other representative methods in nuclear instance segmentation, especially in the context of model generalisability. To achieve automatic nuclear instance segmentation, we propose using a nuclei detection model to provide bounding boxes or central points of nu-clei as visual prompts for SAM in generating nuclear instance masks from histology images.
Uncertainty estimation for out-of-distribution detection in computational histopathology
Oct 18, 2022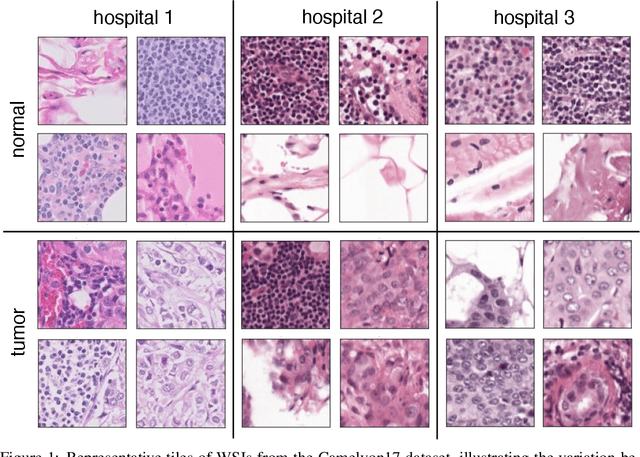
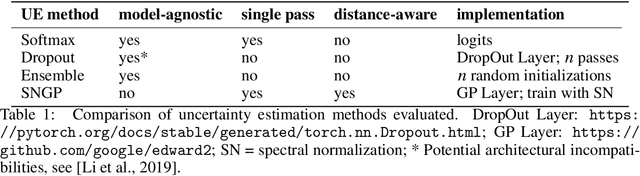
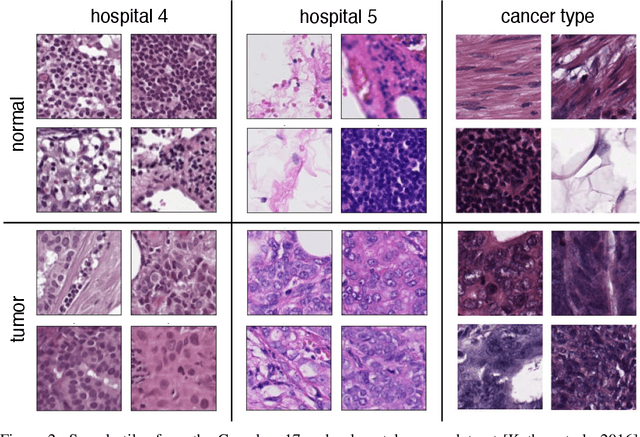
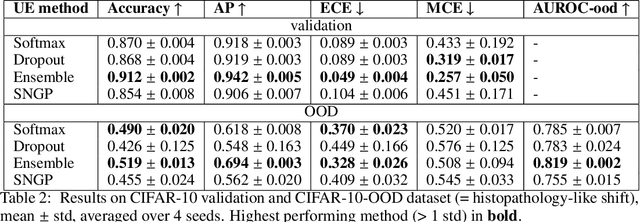
Abstract:In computational histopathology algorithms now outperform humans on a range of tasks, but to date none are employed for automated diagnoses in the clinic. Before algorithms can be involved in such high-stakes decisions they need to "know when they don't know", i.e., they need to estimate their predictive uncertainty. This allows them to defer potentially erroneous predictions to a human pathologist, thus increasing their safety. Here, we evaluate the predictive performance and calibration of several uncertainty estimation methods on clinical histopathology data. We show that a distance-aware uncertainty estimation method outperforms commonly used approaches, such as Monte Carlo dropout and deep ensembles. However, we observe a drop in predictive performance and calibration on novel samples across all uncertainty estimation methods tested. We also investigate the use of uncertainty thresholding to reject out-of-distribution samples for selective prediction. We demonstrate the limitations of this approach and suggest areas for future research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge