Laurent Fanton
Disentangled representations: towards interpretation of sex determination from hip bone
Dec 17, 2021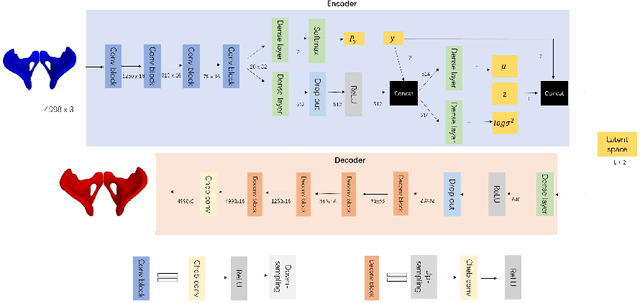

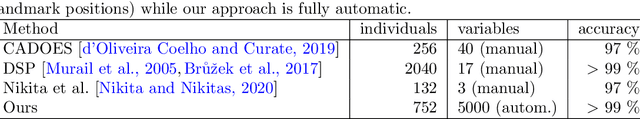
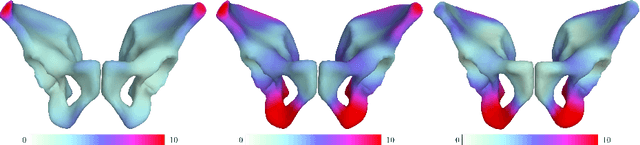
Abstract:By highlighting the regions of the input image that contribute the most to the decision, saliency maps have become a popular method to make neural networks interpretable. In medical imaging, they are particularly well-suited to explain neural networks in the context of abnormality localization. However, from our experiments, they are less suited to classification problems where the features that allow to distinguish between the different classes are spatially correlated, scattered and definitely non-trivial. In this paper we thus propose a new paradigm for better interpretability. To this end we provide the user with relevant and easily interpretable information so that he can form his own opinion. We use Disentangled Variational Auto-Encoders which latent representation is divided into two components: the non-interpretable part and the disentangled part. The latter accounts for the categorical variables explicitly representing the different classes of interest. In addition to providing the class of a given input sample, such a model offers the possibility to transform the sample from a given class to a sample of another class, by modifying the value of the categorical variables in the latent representation. This paves the way to easier interpretation of class differences. We illustrate the relevance of this approach in the context of automatic sex determination from hip bones in forensic medicine. The features encoded by the model, that distinguish the different classes were found to be consistent with expert knowledge.
Hubless keypoint-based 3D deformable groupwise registration
Sep 11, 2018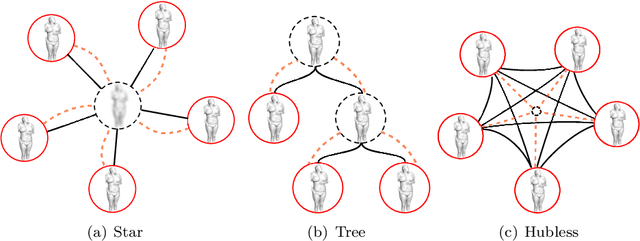
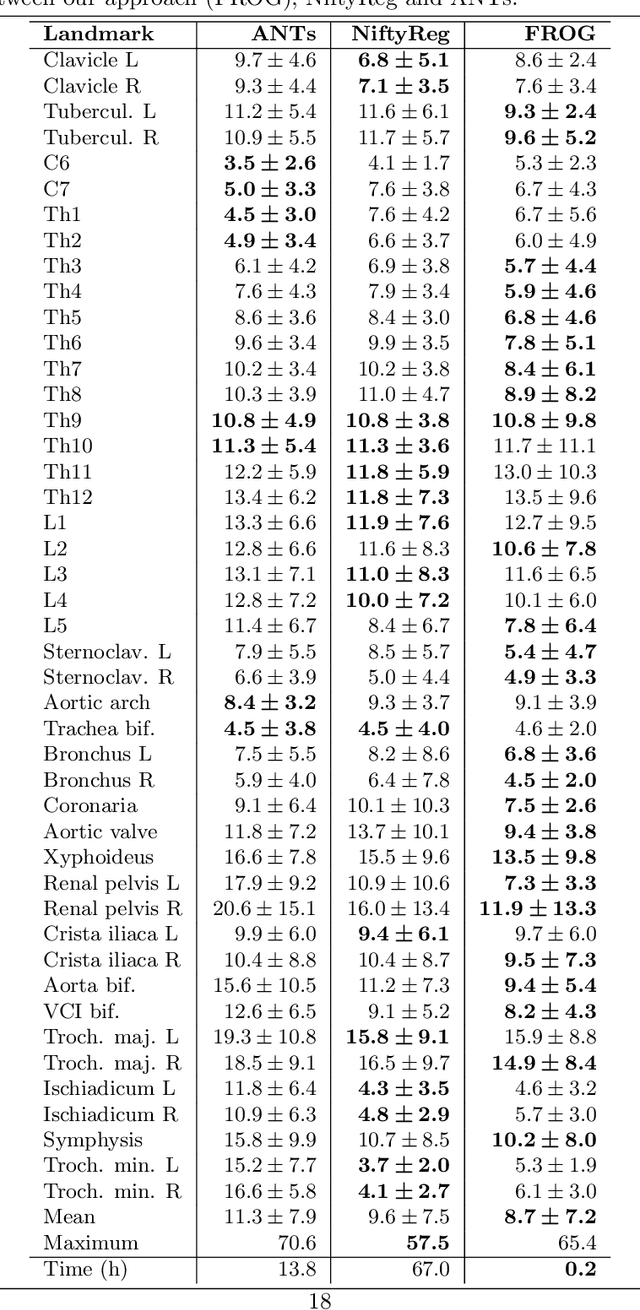
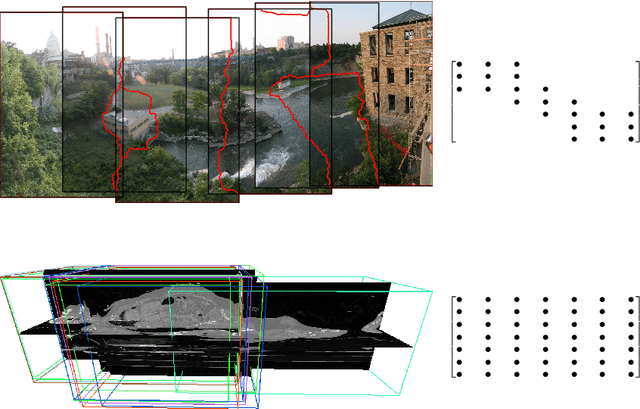
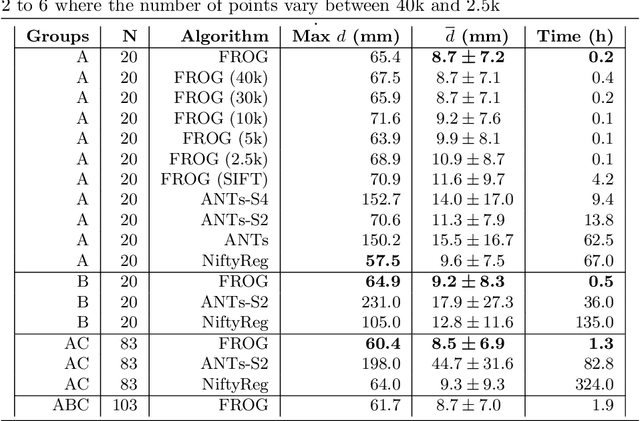
Abstract:We present a novel deformable groupwise registration method, applied to large 3D image groups. Our approach extracts 3D SURF keypoints from images, computes matched pairs of keypoints and registers the group by minimizing pair distances in a hubless way i.e. without computing any central mean image. Using keypoints significantly reduces the problem complexity compared to voxel-based approaches, and enables us to provide an in-core global optimization, similar to the Bundle Adjustment for 3D reconstruction. As we aim at registering images of different patients, the matching step yields many outliers. Then we propose a new EM-weighting algorithm which efficiently discards outliers. Global optimization is carried out with a fast gradient descent algorithm. This allows our approach to robustly register large datasets. The result is a set of half transforms which link the volumes together and can be subsequently exploited for computational anatomy, landmark detection or image segmentation. We show experimental results on whole-body CT scans, with groups of up to 103 volumes. On a benchmark based on anatomical landmarks, our algorithm compares favorably with the star-groupwise voxel-based ANTs and NiftyReg approaches while being much faster. We also discuss the limitations of our approach for lower resolution images such as brain MRI.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge