Kristin Wright-Bettner
Adapting Abstract Meaning Representation Parsing to the Clinical Narrative -- the SPRING THYME parser
May 15, 2024Abstract:This paper is dedicated to the design and evaluation of the first AMR parser tailored for clinical notes. Our objective was to facilitate the precise transformation of the clinical notes into structured AMR expressions, thereby enhancing the interpretability and usability of clinical text data at scale. Leveraging the colon cancer dataset from the Temporal Histories of Your Medical Events (THYME) corpus, we adapted a state-of-the-art AMR parser utilizing continuous training. Our approach incorporates data augmentation techniques to enhance the accuracy of AMR structure predictions. Notably, through this learning strategy, our parser achieved an impressive F1 score of 88% on the THYME corpus's colon cancer dataset. Moreover, our research delved into the efficacy of data required for domain adaptation within the realm of clinical notes, presenting domain adaptation data requirements for AMR parsing. This exploration not only underscores the parser's robust performance but also highlights its potential in facilitating a deeper understanding of clinical narratives through structured semantic representations.
CAMRA: Copilot for AMR Annotation
Nov 18, 2023


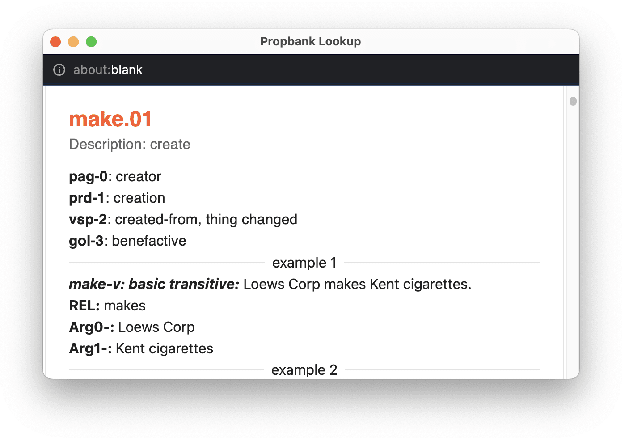
Abstract:In this paper, we introduce CAMRA (Copilot for AMR Annotatations), a cutting-edge web-based tool designed for constructing Abstract Meaning Representation (AMR) from natural language text. CAMRA offers a novel approach to deep lexical semantics annotation such as AMR, treating AMR annotation akin to coding in programming languages. Leveraging the familiarity of programming paradigms, CAMRA encompasses all essential features of existing AMR editors, including example lookup, while going a step further by integrating Propbank roleset lookup as an autocomplete feature within the tool. Notably, CAMRA incorporates AMR parser models as coding co-pilots, greatly enhancing the efficiency and accuracy of AMR annotators. To demonstrate the tool's capabilities, we provide a live demo accessible at: https://camra.colorado.edu
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge