Kim M. Branson
Deep reinforced active learning for multi-class image classification
Jun 20, 2022



Abstract:High accuracy medical image classification can be limited by the costs of acquiring more data as well as the time and expertise needed to label existing images. In this paper, we apply active learning to medical image classification, a method which aims to maximise model performance on a minimal subset from a larger pool of data. We present a new active learning framework, based on deep reinforcement learning, to learn an active learning query strategy to label images based on predictions from a convolutional neural network. Our framework modifies the deep-Q network formulation, allowing us to pick data based additionally on geometric arguments in the latent space of the classifier, allowing for high accuracy multi-class classification in a batch-based active learning setting, enabling the agent to label datapoints that are both diverse and about which it is most uncertain. We apply our framework to two medical imaging datasets and compare with standard query strategies as well as the most recent reinforcement learning based active learning approach for image classification.
Epigenomic language models powered by Cerebras
Dec 14, 2021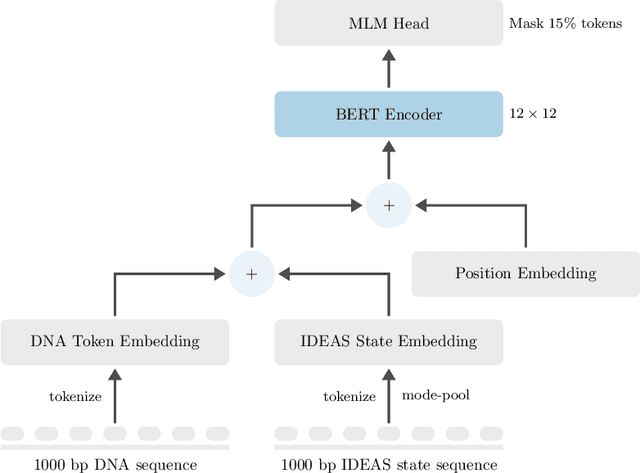
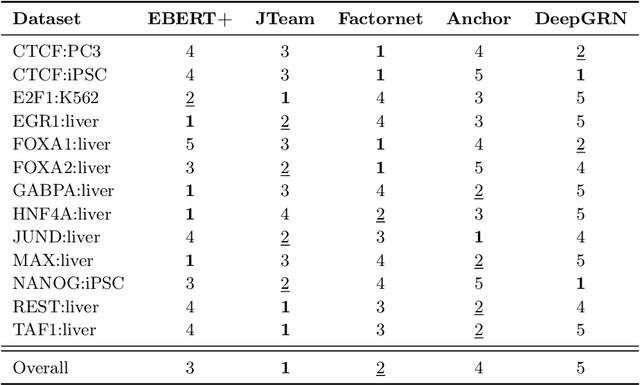
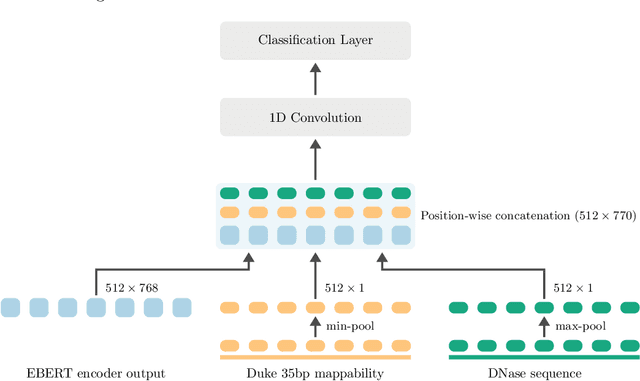
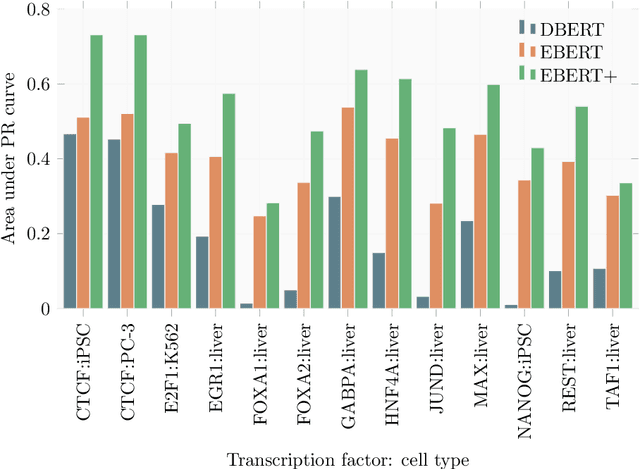
Abstract:Large scale self-supervised pre-training of Transformer language models has advanced the field of Natural Language Processing and shown promise in cross-application to the biological `languages' of proteins and DNA. Learning effective representations of DNA sequences using large genomic sequence corpuses may accelerate the development of models of gene regulation and function through transfer learning. However, to accurately model cell type-specific gene regulation and function, it is necessary to consider not only the information contained in DNA nucleotide sequences, which is mostly invariant between cell types, but also how the local chemical and structural `epigenetic state' of chromosomes varies between cell types. Here, we introduce a Bidirectional Encoder Representations from Transformers (BERT) model that learns representations based on both DNA sequence and paired epigenetic state inputs, which we call Epigenomic BERT (or EBERT). We pre-train EBERT with a masked language model objective across the entire human genome and across 127 cell types. Training this complex model with a previously prohibitively large dataset was made possible for the first time by a partnership with Cerebras Systems, whose CS-1 system powered all pre-training experiments. We show EBERT's transfer learning potential by demonstrating strong performance on a cell type-specific transcription factor binding prediction task. Our fine-tuned model exceeds state of the art performance on 4 of 13 evaluation datasets from ENCODE-DREAM benchmarks and earns an overall rank of 3rd on the challenge leaderboard. We explore how the inclusion of epigenetic data and task specific feature augmentation impact transfer learning performance.
Meta-Learning Initializations for Low-Resource Drug Discovery
Mar 12, 2020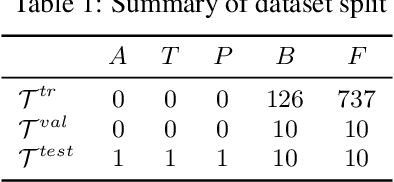
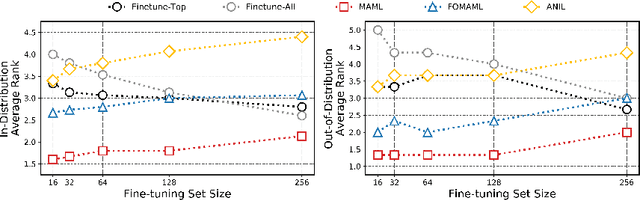
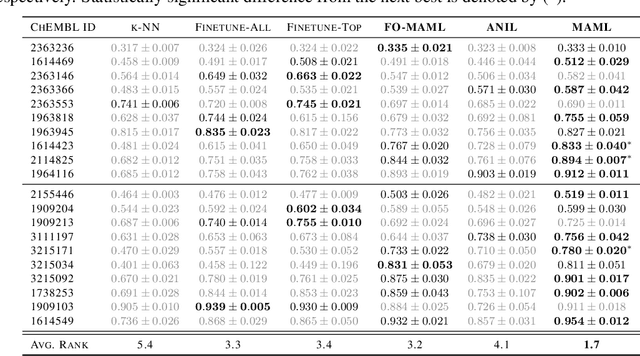
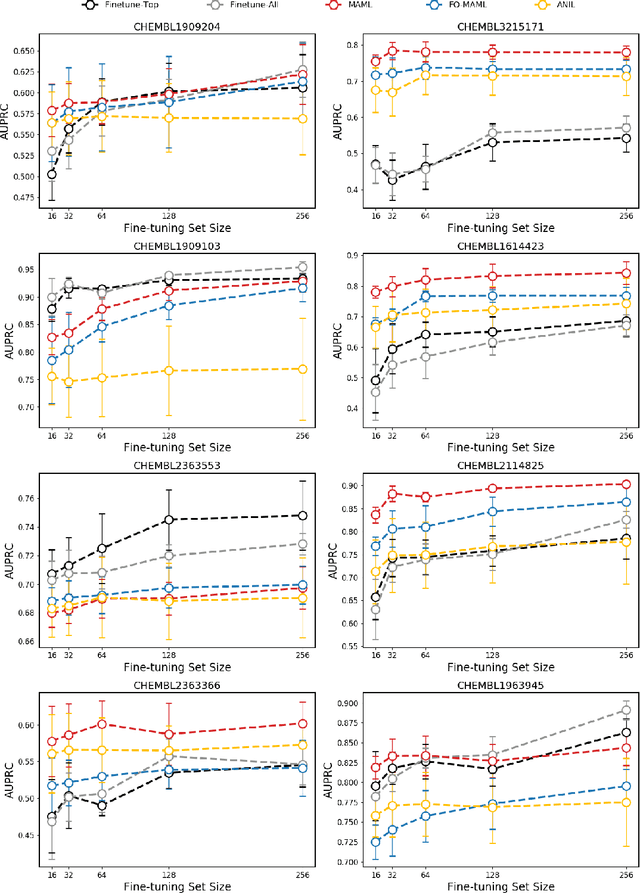
Abstract:Building in silico models to predict chemical properties and activities is a crucial step in drug discovery. However, drug discovery projects are often characterized by limited labeled data, hindering the applications of deep learning in this setting. Meanwhile advances in meta-learning have enabled state-of-the-art performances in few-shot learning benchmarks, naturally prompting the question: Can meta-learning improve deep learning performance in low-resource drug discovery projects? In this work, we assess the efficiency of the Model-Agnostic Meta-Learning (MAML) algorithm - along with its variants FO-MAML and ANIL - at learning to predict chemical properties and activities. Using the ChEMBL20 dataset to emulate low-resource settings, our benchmark shows that meta-initializations perform comparably to or outperform multi-task pre-training baselines on 16 out of 20 in-distribution tasks and on all out-of-distribution tasks, providing an average improvement in AUPRC of 7.2% and 14.9% respectively. Finally, we observe that meta-initializations consistently result in the best performing models across fine-tuning sets with $k \in \{16, 32, 64, 128, 256\}$ instances.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge