Kevin J. Parker
Difference Autocorrelation: A Novel Approach to Estimate Shear Wave Speed in the Presence of Compression Waves
Jun 18, 2024



Abstract:In share wave elastography (SWE), the aim is to measure the velocity of shear waves, however unwanted compression waves and bulk tissue motion pose challenges in evaluating tissue stiffness. Conventional approaches often struggle to discriminate between shear and compression waves, leading to inaccurate shear wave speed (SWS) estimation. In this study, we propose a novel approach known as the difference autocorrelation estimator to accurately estimate reverberant SWS in the presence of compression waves and noise. Methods: The difference autocorrelation estimator, unlike conventional techniques, computes the subtraction of velocity between neighboring particles, effectively minimizing the impact of long wavelength compression waves and other wide-area movements such as those caused by respiration. We evaluated the effectiveness of the integrated difference autocorrelation (IDA) by: (1) using k-Wave simulations of a branching cylinder in a soft background, (2) using ultrasound elastography on a breast phantom, (3) using ultrasound elastography in the human liver-kidney region, and (4) using magnetic resonance elastography (MRE) on a brain phantom. Results: By applying IDA on the unfiltered contaminated wave fields of simulation and elastography experiments, the estimated SWSs are in good agreement with the ground truth values (i.e., less than 2% error for the simulation, 9% error for ultrasound elastography of breast phantom and 19% error for MRE). Conclusion: Our results demonstrate that IDA accurately estimates SWS, revealing the existence of a lesion, even in the presence of strong compression waves. Significance: IDA exhibits consistency in SWS estimation across different modalities and excitation scenarios, highlighting its robustness and potential clinical utility.
Multiparametric quantification and visualization of liver fat using ultrasound
Apr 02, 2024



Abstract:Objectives- Several ultrasound measures have shown promise for assessment of steatosis compared to traditional B-scan, however clinicians may be required to integrate information across the parameters. Here, we propose an integrated multiparametric approach, enabling simple clinical assessment of key information from combined ultrasound parameters. Methods- We have measured 13 parameters related to ultrasound and shear wave elastography. These were measured in 30 human subjects under a study of liver fat. The 13 individual measures are assessed for their predictive value using independent magnetic resonance imaging-derived proton density fat fraction (MRI-PDFF) measurements as a reference standard. In addition, a comprehensive and fine-grain analysis is made of all possible combinations of sub-sets of these parameters to determine if any subset can be efficiently combined to predict fat fraction. Results- We found that as few as four key parameters related to ultrasound propagation are sufficient to generate a linear multiparametric parameter with a correlation against MRI-PDFF values of greater than 0.93. This optimal combination was found to have a classification area under the curve (AUC) approaching 1.0 when applying a threshold for separating steatosis grade zero from higher classes. Furthermore, a strategy is developed for applying local estimates of fat content as a color overlay to produce a visual impression of the extent and distribution of fat within the liver. Conclusion- In principle, this approach can be applied to most clinical ultrasound systems to provide the clinician and patient with a rapid and inexpensive estimate of liver fat content.
WATUNet: A Deep Neural Network for Segmentation of Volumetric Sweep Imaging Ultrasound
Nov 17, 2023Abstract:Objective. Limited access to breast cancer diagnosis globally leads to delayed treatment. Ultrasound, an effective yet underutilized method, requires specialized training for sonographers, which hinders its widespread use. Approach. Volume sweep imaging (VSI) is an innovative approach that enables untrained operators to capture high-quality ultrasound images. Combined with deep learning, like convolutional neural networks (CNNs), it can potentially transform breast cancer diagnosis, enhancing accuracy, saving time and costs, and improving patient outcomes. The widely used UNet architecture, known for medical image segmentation, has limitations, such as vanishing gradients and a lack of multi-scale feature extraction and selective region attention. In this study, we present a novel segmentation model known as Wavelet_Attention_UNet (WATUNet). In this model, we incorporate wavelet gates (WGs) and attention gates (AGs) between the encoder and decoder instead of a simple connection to overcome the limitations mentioned, thereby improving model performance. Main results. Two datasets are utilized for the analysis. The public "Breast Ultrasound Images" (BUSI) dataset of 780 images and a VSI dataset of 3818 images. Both datasets contained segmented lesions categorized into three types: no mass, benign mass, and malignant mass. Our segmentation results show superior performance compared to other deep networks. The proposed algorithm attained a Dice coefficient of 0.94 and an F1 score of 0.94 on the VSI dataset and scored 0.93 and 0.94 on the public dataset, respectively.
Limitations of curl and directional filters in elastography
May 15, 2023Abstract:In the approaches to elastography, two mathematical operations have been frequently applied to improve the final estimate of shear wave speed and shear modulus of tissues. The vector curl operator can separate out the transverse component of a complicated displacement field, and directional filters can separate distinct orientations of wave propagation. However, there are practical limitations that can prevent the intended improvement in elastography estimates. Some simple configurations of wavefields relevant to elastography are examined against theoretical models within the semi-infinite elastic medium and guided waves in a bounded medium. The Miller-Pursey solutions in simplified form are examined for the semi-infinite medium and the Lamb wave symmetric form is considered for the guided wave structure. In both cases, wave combinations along with practical limits on the imaging plane can prevent the curl and directional filter operations from directly providing an improved measure of shear wave speed and shear modulus. Additional limits on signal-to-noise and the support of filters also restrict the applica-bility of these strategies for improving elastographic measures. Practical implementations of shear wave excitations applied to the body and to bounded structures within the body can involve waves that are not easily resolved by the vector curl operator and directional filters. These limits may be overcome by more advanced strategies or simple improvements in baseline parameters including the size of the region of interest and the number of shear waves propagated within.
Improving the diagnosis of breast cancer based on biophysical ultrasound features utilizing machine learning
Jul 13, 2022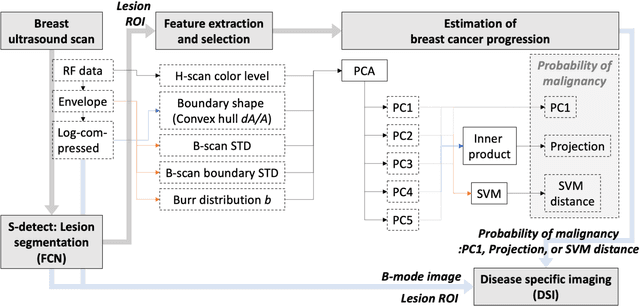

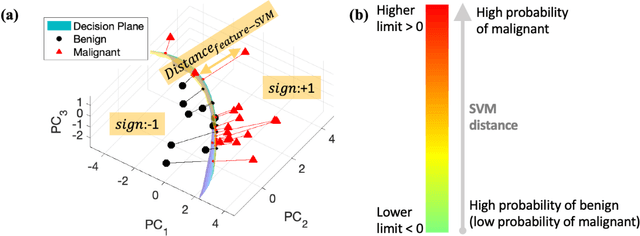
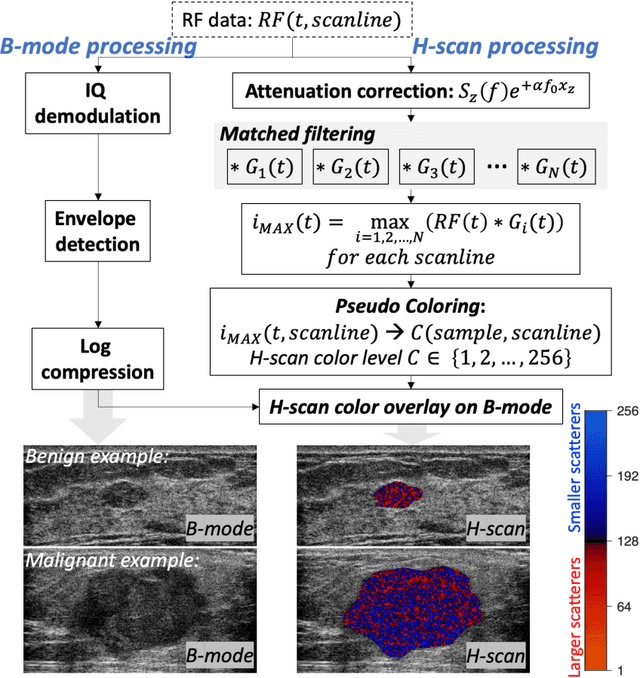
Abstract:The improved diagnostic accuracy of ultrasound breast examinations remains an important goal. In this study, we propose a biophysical feature based machine learning method for breast cancer detection to improve the performance beyond a benchmark deep learning algorithm and to furthermore provide a color overlay visual map of the probability of malignancy within a lesion. This overall framework is termed disease specific imaging. Previously, 150 breast lesions were segmented and classified utilizing a modified fully convolutional network and a modified GoogLeNet, respectively. In this study multiparametric analysis was performed within the contoured lesions. Features were extracted from ultrasound radiofrequency, envelope, and log compressed data based on biophysical and morphological models. The support vector machine with a Gaussian kernel constructed a nonlinear hyperplane, and we calculated the distance between the hyperplane and data point of each feature in multiparametric space. The distance can quantitatively assess a lesion, and suggest the probability of malignancy that is color coded and overlaid onto B mode images. Training and evaluation were performed on in vivo patient data. The overall accuracy for the most common types and sizes of breast lesions in our study exceeded 98.0% for classification and 0.98 for an area under the receiver operating characteristic curve, which is more precise than the performance of radiologists and a deep learning system. Further, the correlation between the probability and BI RADS enables a quantitative guideline to predict breast cancer. Therefore, we anticipate that the proposed framework can help radiologists achieve more accurate and convenient breast cancer classification and detection.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge