Keilung Choy
A Symbolic and Statistical Learning Framework to Discover Bioprocessing Regulatory Mechanism: Cell Culture Example
May 06, 2025
Abstract:Bioprocess mechanistic modeling is essential for advancing intelligent digital twin representation of biomanufacturing, yet challenges persist due to complex intracellular regulation, stochastic system behavior, and limited experimental data. This paper introduces a symbolic and statistical learning framework to identify key regulatory mechanisms and quantify model uncertainty. Bioprocess dynamics is formulated with stochastic differential equations characterizing intrinsic process variability, with a predefined set of candidate regulatory mechanisms constructed from biological knowledge. A Bayesian learning approach is developed, which is based on a joint learning of kinetic parameters and regulatory structure through a formulation of the mixture model. To enhance computational efficiency, a Metropolis-adjusted Langevin algorithm with adjoint sensitivity analysis is developed for posterior exploration. Compared to state-of-the-art Bayesian inference approaches, the proposed framework achieves improved sample efficiency and robust model selection. An empirical study demonstrates its ability to recover missing regulatory mechanisms and improve model fidelity under data-limited conditions.
Digital Twin Calibration with Model-Based Reinforcement Learning
Jan 04, 2025Abstract:This paper presents a novel methodological framework, called the Actor-Simulator, that incorporates the calibration of digital twins into model-based reinforcement learning for more effective control of stochastic systems with complex nonlinear dynamics. Traditional model-based control often relies on restrictive structural assumptions (such as linear state transitions) and fails to account for parameter uncertainty in the model. These issues become particularly critical in industries such as biopharmaceutical manufacturing, where process dynamics are complex and not fully known, and only a limited amount of data is available. Our approach jointly calibrates the digital twin and searches for an optimal control policy, thus accounting for and reducing model error. We balance exploration and exploitation by using policy performance as a guide for data collection. This dual-component approach provably converges to the optimal policy, and outperforms existing methods in extensive numerical experiments based on the biopharmaceutical manufacturing domain.
Adjoint Sensitivity Analysis on Multi-Scale Bioprocess Stochastic Reaction Network
May 07, 2024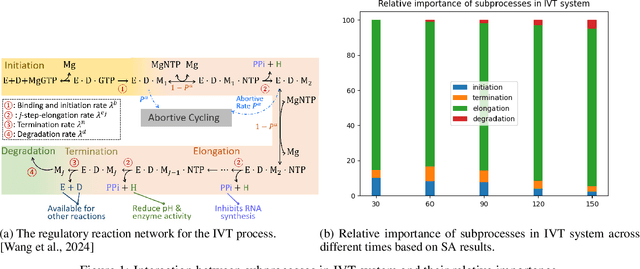
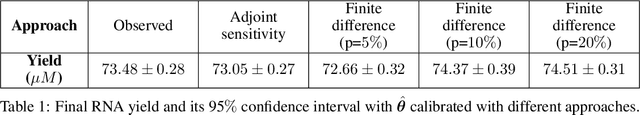
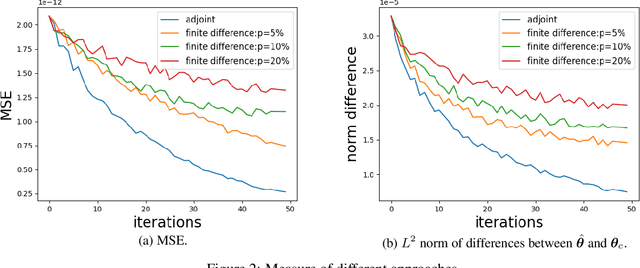
Abstract:Motivated by the pressing challenges in the digital twin development for biomanufacturing process, we introduce an adjoint sensitivity analysis (SA) approach to expedite the learning of mechanistic model parameters. In this paper, we consider enzymatic stochastic reaction networks representing a multi-scale bioprocess mechanistic model that allows us to integrate disparate data from diverse production processes and leverage the information from existing macro-kinetic and genome-scale models. To support forward prediction and backward reasoning, we develop a convergent adjoint SA algorithm studying how the perturbations of model parameters and inputs (e.g., initial state) propagate through enzymatic reaction networks and impact on output trajectory predictions. This SA can provide a sample efficient and interpretable way to assess the sensitivities between inputs and outputs accounting for their causal dependencies. Our empirical study underscores the resilience of these sensitivities and illuminates a deeper comprehension of the regulatory mechanisms behind bioprocess through sensitivities.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge