Kaupo Palo
Self-Supervised Single-Image Deconvolution with Siamese Neural Networks
Aug 18, 2023



Abstract:Inverse problems in image reconstruction are fundamentally complicated by unknown noise properties. Classical iterative deconvolution approaches amplify noise and require careful parameter selection for an optimal trade-off between sharpness and grain. Deep learning methods allow for flexible parametrization of the noise and learning its properties directly from the data. Recently, self-supervised blind-spot neural networks were successfully adopted for image deconvolution by including a known point-spread function in the end-to-end training. However, their practical application has been limited to 2D images in the biomedical domain because it implies large kernels that are poorly optimized. We tackle this problem with Fast Fourier Transform convolutions that provide training speed-up in 3D microscopy deconvolution tasks. Further, we propose to adopt a Siamese invariance loss for deconvolution and empirically identify its optimal position in the neural network between blind-spot and full image branches. The experimental results show that our improved framework outperforms the previous state-of-the-art deconvolution methods with a known point spread function.
Metadata Improves Segmentation Through Multitasking Elicitation
Aug 18, 2023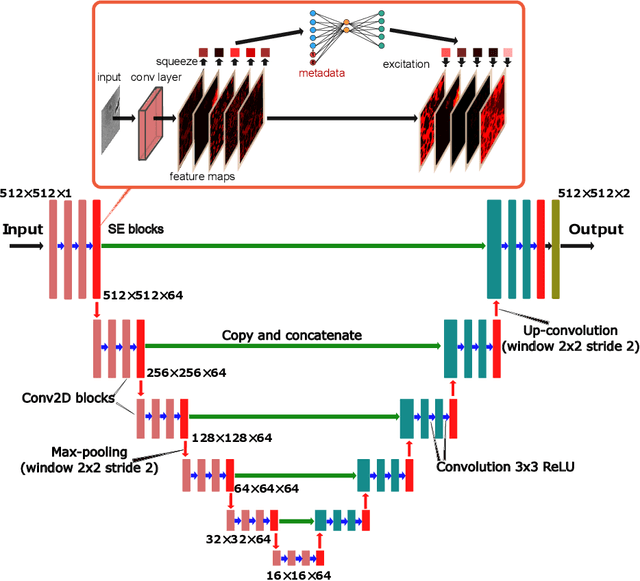
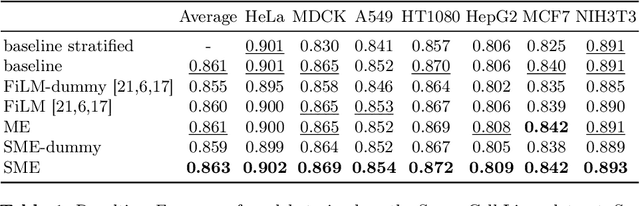
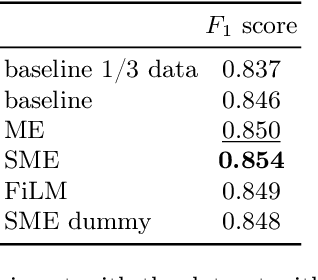
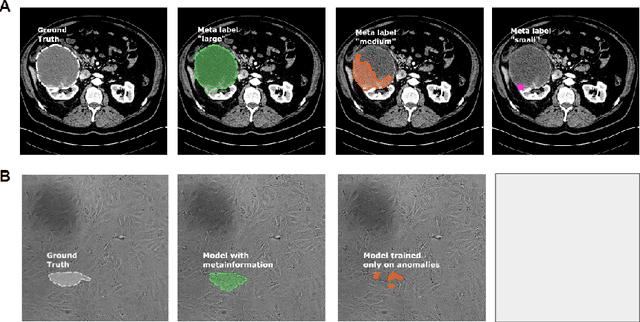
Abstract:Metainformation is a common companion to biomedical images. However, this potentially powerful additional source of signal from image acquisition has had limited use in deep learning methods, for semantic segmentation in particular. Here, we incorporate metadata by employing a channel modulation mechanism in convolutional networks and study its effect on semantic segmentation tasks. We demonstrate that metadata as additional input to a convolutional network can improve segmentation results while being inexpensive in implementation as a nimble add-on to popular models. We hypothesize that this benefit of metadata can be attributed to facilitating multitask switching. This aspect of metadata-driven systems is explored and discussed in detail.
Noise2Stack: Improving Image Restoration by Learning from Volumetric Data
Nov 10, 2020
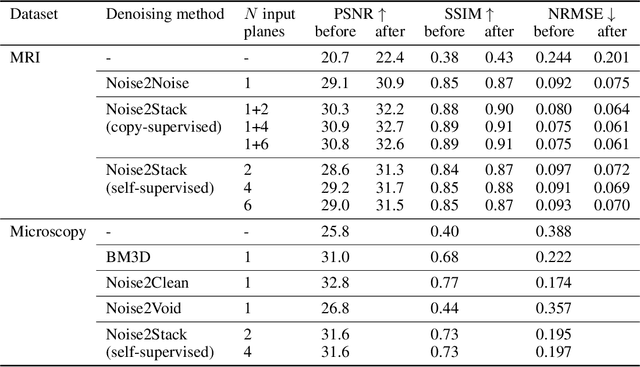
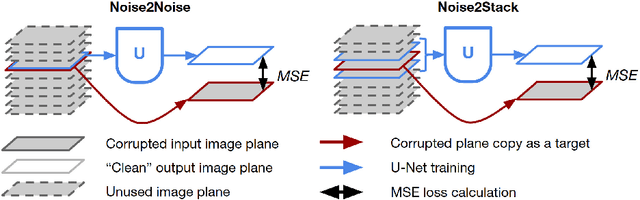
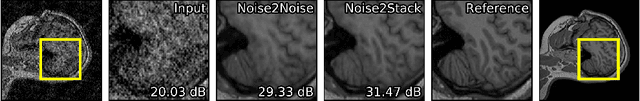
Abstract:Biomedical images are noisy. The imaging equipment itself has physical limitations, and the consequent experimental trade-offs between signal-to-noise ratio, acquisition speed, and imaging depth exacerbate the problem. Denoising is, therefore, an essential part of any image processing pipeline, and convolutional neural networks are currently the method of choice for this task. One popular approach, Noise2Noise, does not require clean ground truth, and instead, uses a second noisy copy as a training target. Self-supervised methods, like Noise2Self and Noise2Void, relax data requirements by learning the signal without an explicit target but are limited by the lack of information in a single image. Here, we introduce Noise2Stack, an extension of the Noise2Noise method to image stacks that takes advantage of a shared signal between spatially neighboring planes. Our experiments on magnetic resonance brain scans and newly acquired multiplane microscopy data show that learning only from image neighbors in a stack is sufficient to outperform Noise2Noise and Noise2Void and close the gap to supervised denoising methods. Our findings point towards low-cost, high-reward improvement in the denoising pipeline of multiplane biomedical images. As a part of this work, we release a microscopy dataset to establish a benchmark for the multiplane image denoising.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge