Justin Wood
Parallel development of social preferences in fish and machines
May 18, 2023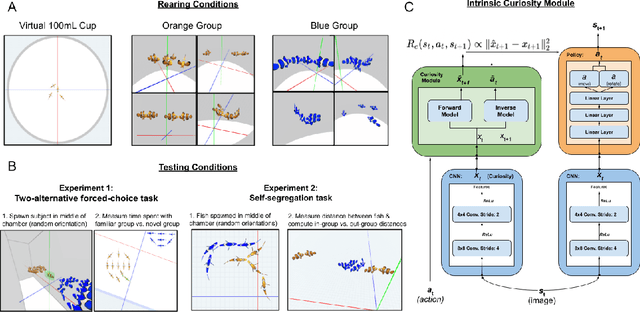
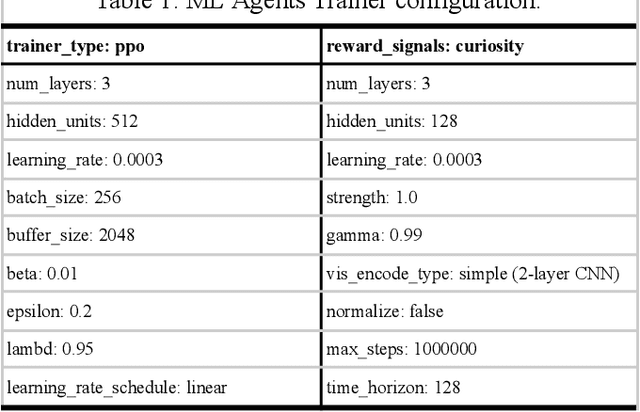
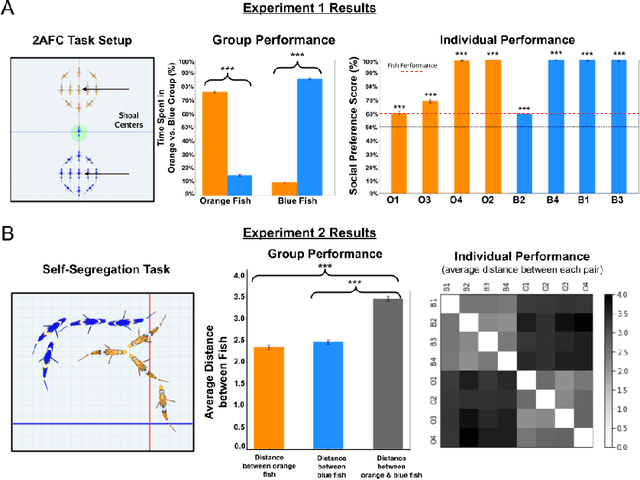
Abstract:What are the computational foundations of social grouping? Traditional approaches to this question have focused on verbal reasoning or simple (low-dimensional) quantitative models. In the real world, however, social preferences emerge when high-dimensional learning systems (brains and bodies) interact with high-dimensional sensory inputs during an animal's embodied interactions with the world. A deep understanding of social grouping will therefore require embodied models that learn directly from sensory inputs using high-dimensional learning mechanisms. To this end, we built artificial neural networks (ANNs), embodied those ANNs in virtual fish bodies, and raised the artificial fish in virtual fish tanks that mimicked the rearing conditions of real fish. We then compared the social preferences that emerged in real fish versus artificial fish. We found that when artificial fish had two core learning mechanisms (reinforcement learning and curiosity-driven learning), artificial fish developed fish-like social preferences. Like real fish, the artificial fish spontaneously learned to prefer members of their own group over members of other groups. The artificial fish also spontaneously learned to self-segregate with their in-group, akin to self-segregation behavior seen in nature. Our results suggest that social grouping can emerge from three ingredients: (1) reinforcement learning, (2) intrinsic motivation, and (3) early social experiences with in-group members. This approach lays a foundation for reverse engineering animal-like social behavior with image-computable models, bridging the divide between high-dimensional sensory inputs and social preferences.
OpBerg: Discovering causal sentences using optimal alignments
Apr 03, 2019Abstract:The biological literature is rich with sentences that describe causal relations. Methods that automatically extract such sentences can help biologists to synthesize the literature and even discover latent relations that had not been articulated explicitly. Current methods for extracting causal sentences are based on either machine learning or a predefined database of causal terms. Machine learning approaches require a large set of labeled training data and can be susceptible to noise. Methods based on predefined databases are limited by the quality of their curation and are unable to capture new concepts or mistakes in the input. We address these challenges by adapting and improving a method designed for a seemingly unrelated problem: finding alignments between genomic sequences. This paper presents a novel and outperforming method for extracting causal relations from text by aligning the part-of-speech representations of an input set with that of known causal sentences. Our experiments show that when applied to the task of finding causal sentences in biological literature, our method improves on the accuracy of other methods in a computationally efficient manner.
Source-LDA: Enhancing probabilistic topic models using prior knowledge sources
May 17, 2017



Abstract:A popular approach to topic modeling involves extracting co-occurring n-grams of a corpus into semantic themes. The set of n-grams in a theme represents an underlying topic, but most topic modeling approaches are not able to label these sets of words with a single n-gram. Such labels are useful for topic identification in summarization systems. This paper introduces a novel approach to labeling a group of n-grams comprising an individual topic. The approach taken is to complement the existing topic distributions over words with a known distribution based on a predefined set of topics. This is done by integrating existing labeled knowledge sources representing known potential topics into the probabilistic topic model. These knowledge sources are translated into a distribution and used to set the hyperparameters of the Dirichlet generated distribution over words. In the inference these modified distributions guide the convergence of the latent topics to conform with the complementary distributions. This approach ensures that the topic inference process is consistent with existing knowledge. The label assignment from the complementary knowledge sources are then transferred to the latent topics of the corpus. The results show both accurate label assignment to topics as well as improved topic generation than those obtained using various labeling approaches based off Latent Dirichlet allocation (LDA).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge