Julien Hogan
Noise Pollution in Hospital Readmission Prediction: Long Document Classification with Reinforcement Learning
May 23, 2020



Abstract:This paper presents a reinforcement learning approach to extract noise in long clinical documents for the task of readmission prediction after kidney transplant. We face the challenges of developing robust models on a small dataset where each document may consist of over 10K tokens with full of noise including tabular text and task-irrelevant sentences. We first experiment four types of encoders to empirically decide the best document representation, and then apply reinforcement learning to remove noisy text from the long documents, which models the noise extraction process as a sequential decision problem. Our results show that the old bag-of-words encoder outperforms deep learning-based encoders on this task, and reinforcement learning is able to improve upon baseline while pruning out 25% text segments. Our analysis depicts that reinforcement learning is able to identify both typical noisy tokens and task-specific noisy text.
Multimodal Ensemble Approach to Incorporate Various Types of Clinical Notes for Predicting Readmission
May 31, 2019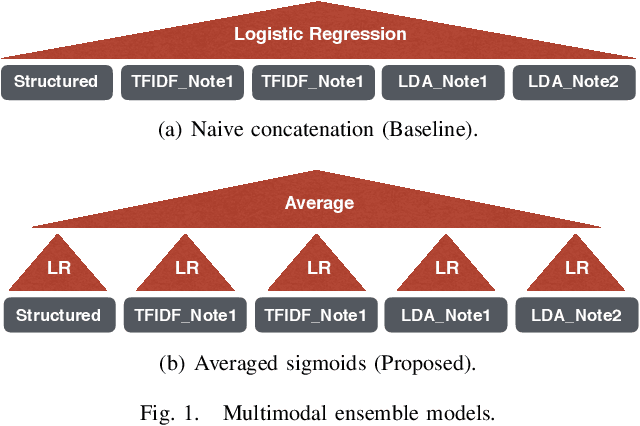
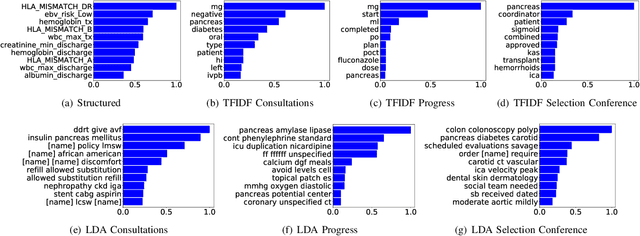
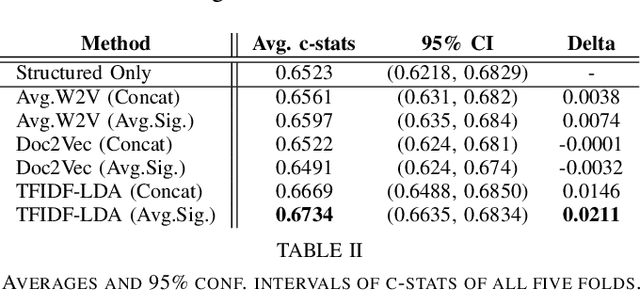
Abstract:Electronic Health Records (EHRs) have been heavily used to predict various downstream clinical tasks such as readmission or mortality. One of the modalities in EHRs, clinical notes, has not been fully explored for these tasks due to its unstructured and inexplicable nature. Although recent advances in deep learning (DL) enables models to extract interpretable features from unstructured data, they often require a large amount of training data. However, many tasks in medical domains inherently consist of small sample data with lengthy documents; for a kidney transplant as an example, data from only a few thousand of patients are available and each patient's document consists of a couple of millions of words in major hospitals. Thus, complex DL methods cannot be applied to these kinds of domains. In this paper, we present a comprehensive ensemble model using vector space modeling and topic modeling. Our proposed model is evaluated on the readmission task of kidney transplant patients and improves 0.0211 in terms of c-statistics from the previous state-of-the-art approach using structured data, while typical DL methods fail to beat this approach. The proposed architecture provides the interpretable score for each feature from both modalities, structured and unstructured data, which is shown to be meaningful through a physician's evaluation.
* 4 pages, IEEE BHI 2019
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge