Jon Braatz
Deep Learning-Based Sparse Whole-Slide Image Analysis for the Diagnosis of Gastric Intestinal Metaplasia
Jan 05, 2022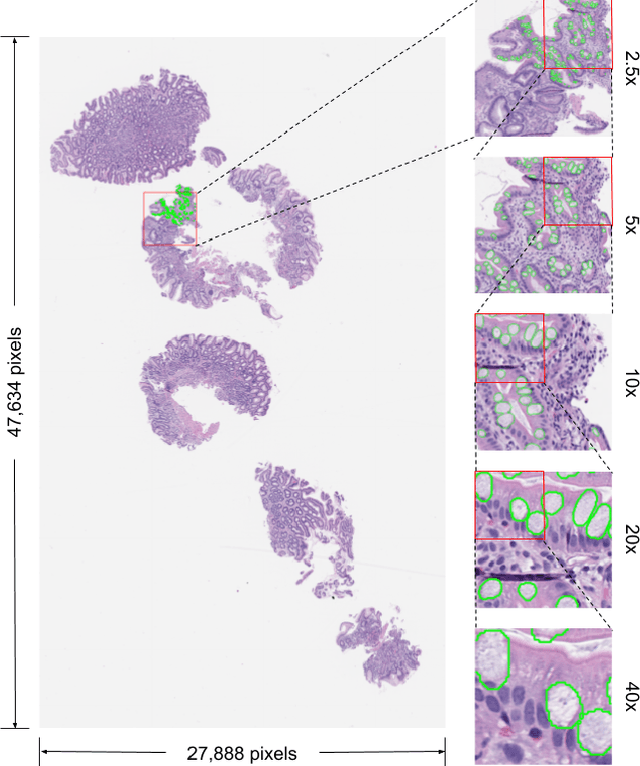
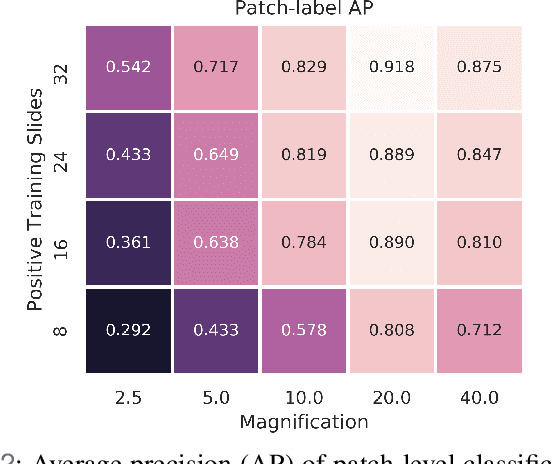

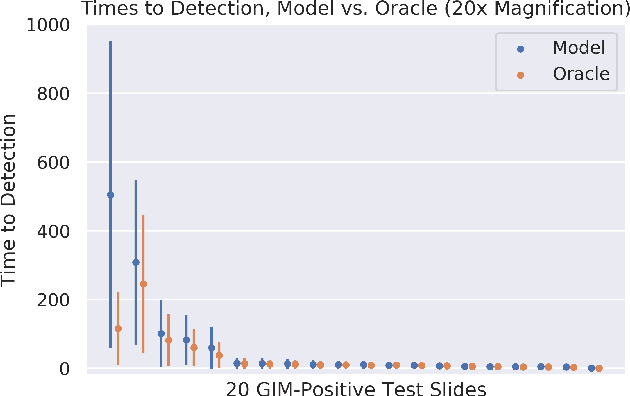
Abstract:In recent years, deep learning has successfully been applied to automate a wide variety of tasks in diagnostic histopathology. However, fast and reliable localization of small-scale regions-of-interest (ROI) has remained a key challenge, as discriminative morphologic features often occupy only a small fraction of a gigapixel-scale whole-slide image (WSI). In this paper, we propose a sparse WSI analysis method for the rapid identification of high-power ROI for WSI-level classification. We develop an evaluation framework inspired by the early classification literature, in order to quantify the tradeoff between diagnostic performance and inference time for sparse analytic approaches. We test our method on a common but time-consuming task in pathology - that of diagnosing gastric intestinal metaplasia (GIM) on hematoxylin and eosin (H&E)-stained slides from endoscopic biopsy specimens. GIM is a well-known precursor lesion along the pathway to development of gastric cancer. We performed a thorough evaluation of the performance and inference time of our approach on a test set of GIM-positive and GIM-negative WSI, finding that our method successfully detects GIM in all positive WSI, with a WSI-level classification area under the receiver operating characteristic curve (AUC) of 0.98 and an average precision (AP) of 0.95. Furthermore, we show that our method can attain these metrics in under one minute on a standard CPU. Our results are applicable toward the goal of developing neural networks that can easily be deployed in clinical settings to support pathologists in quickly localizing and diagnosing small-scale morphologic features in WSI.
Selecting Regions of Interest in Large Multi-Scale Images for Cancer Pathology
Jul 03, 2020
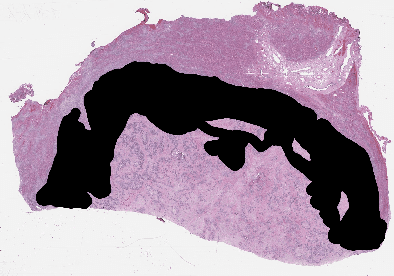
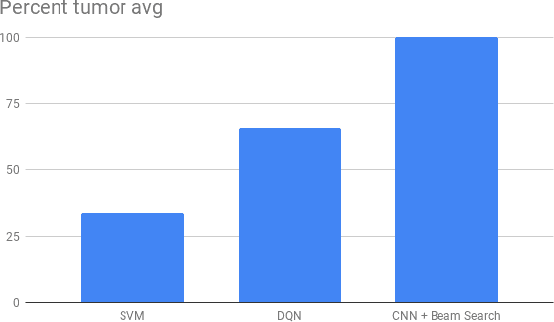
Abstract:Recent breakthroughs in object detection and image classification using Convolutional Neural Networks (CNNs) are revolutionizing the state of the art in medical imaging, and microscopy in particular presents abundant opportunities for computer vision algorithms to assist medical professionals in diagnosis of diseases ranging from malaria to cancer. High resolution scans of microscopy slides called Whole Slide Images (WSIs) offer enough information for a cancer pathologist to come to a conclusion regarding cancer presence, subtype, and severity based on measurements of features within the slide image at multiple scales and resolutions. WSIs' extremely high resolutions and feature scales ranging from gross anatomical structures down to cell nuclei preclude the use of standard CNN models for object detection and classification, which have typically been designed for images with dimensions in the hundreds of pixels and with objects on the order of the size of the image itself. We explore parallel approaches based on Reinforcement Learning and Beam Search to learn to progressively zoom into the WSI to detect Regions of Interest (ROIs) in liver pathology slides containing one of two types of liver cancer, namely Hepatocellular Carcinoma (HCC) and Cholangiocarcinoma (CC). These ROIs can then be presented directly to the pathologist to aid in measurement and diagnosis or be used for automated classification of tumor subtype.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge