Jitender Singh Virk
MRI to PET Cross-Modality Translation using Globally and Locally Aware GAN for Multi-Modal Diagnosis of Alzheimer's Disease
Aug 04, 2021



Abstract:Medical imaging datasets are inherently high dimensional with large variability and low sample sizes that limit the effectiveness of deep learning algorithms. Recently, generative adversarial networks (GANs) with the ability to synthesize realist images have shown great potential as an alternative to standard data augmentation techniques. Our work focuses on cross-modality synthesis of fluorodeoxyglucose~(FDG) Positron Emission Tomography~(PET) scans from structural Magnetic Resonance~(MR) images using generative models to facilitate multi-modal diagnosis of Alzheimer's disease (AD). Specifically, we propose a novel end-to-end, globally and locally aware image-to-image translation GAN (GLA-GAN) with a multi-path architecture that enforces both global structural integrity and fidelity to local details. We further supplement the standard adversarial loss with voxel-level intensity, multi-scale structural similarity (MS-SSIM) and region-of-interest (ROI) based loss components that reduce reconstruction error, enforce structural consistency at different scales and perceive variation in regional sensitivity to AD respectively. Experimental results demonstrate that our GLA-GAN not only generates synthesized FDG-PET scans with enhanced image quality but also superior clinical utility in improving AD diagnosis compared to state-of-the-art models. Finally, we attempt to interpret some of the internal units of the GAN that are closely related to this specific cross-modality generation task.
Domain Specific, Semi-Supervised Transfer Learning for Medical Imaging
May 24, 2020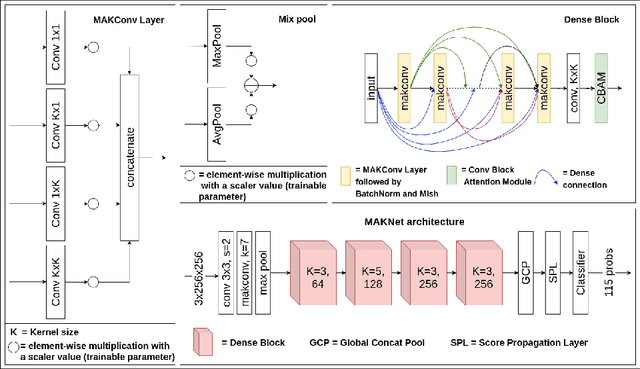
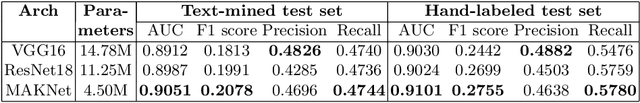
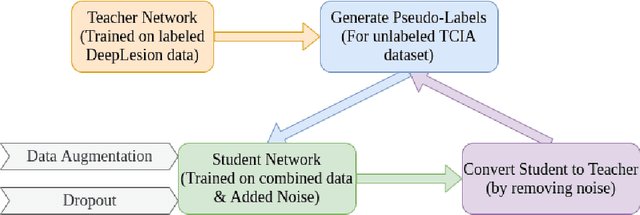

Abstract:Limited availability of annotated medical imaging data poses a challenge for deep learning algorithms. Although transfer learning minimizes this hurdle in general, knowledge transfer across disparate domains is shown to be less effective. On the other hand, smaller architectures were found to be more compelling in learning better features. Consequently, we propose a lightweight architecture that uses mixed asymmetric kernels (MAKNet) to reduce the number of parameters significantly. Additionally, we train the proposed architecture using semi-supervised learning to provide pseudo-labels for a large medical dataset to assist with transfer learning. The proposed MAKNet provides better classification performance with $60 - 70\%$ less parameters than popular architectures. Experimental results also highlight the importance of domain-specific knowledge for effective transfer learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge