Ji Young Lee
PubMed 200k RCT: a Dataset for Sequential Sentence Classification in Medical Abstracts
Oct 17, 2017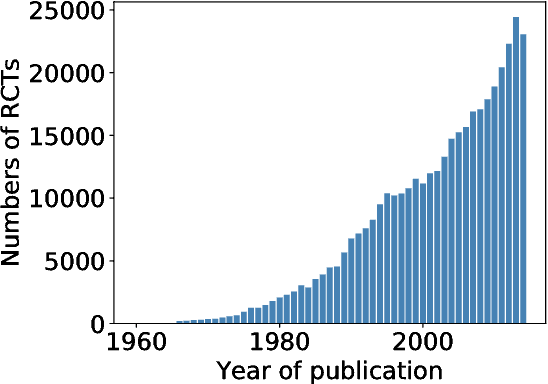
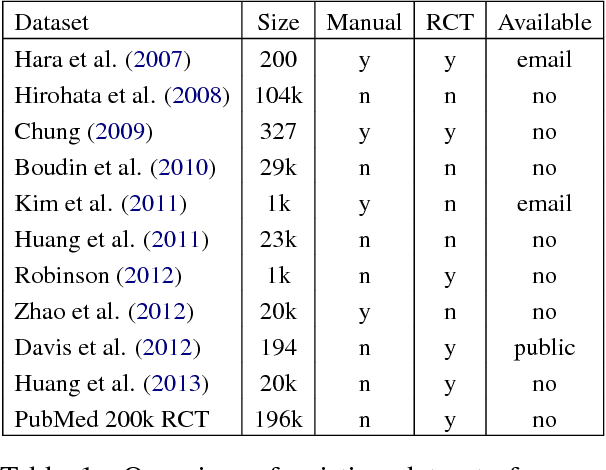
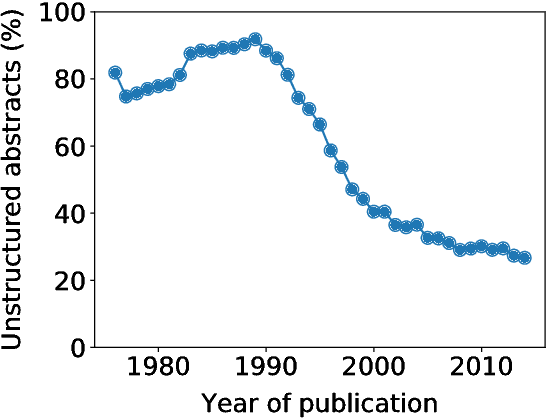
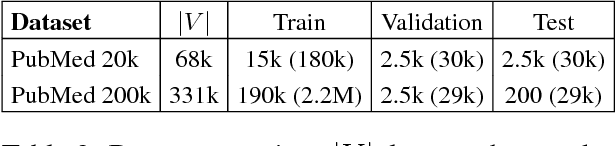
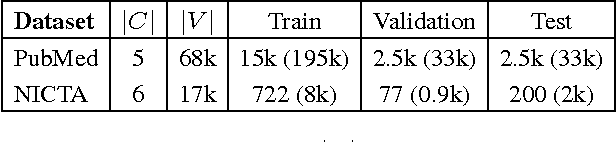
Abstract:We present PubMed 200k RCT, a new dataset based on PubMed for sequential sentence classification. The dataset consists of approximately 200,000 abstracts of randomized controlled trials, totaling 2.3 million sentences. Each sentence of each abstract is labeled with their role in the abstract using one of the following classes: background, objective, method, result, or conclusion. The purpose of releasing this dataset is twofold. First, the majority of datasets for sequential short-text classification (i.e., classification of short texts that appear in sequences) are small: we hope that releasing a new large dataset will help develop more accurate algorithms for this task. Second, from an application perspective, researchers need better tools to efficiently skim through the literature. Automatically classifying each sentence in an abstract would help researchers read abstracts more efficiently, especially in fields where abstracts may be long, such as the medical field.
Transfer Learning for Named-Entity Recognition with Neural Networks
May 17, 2017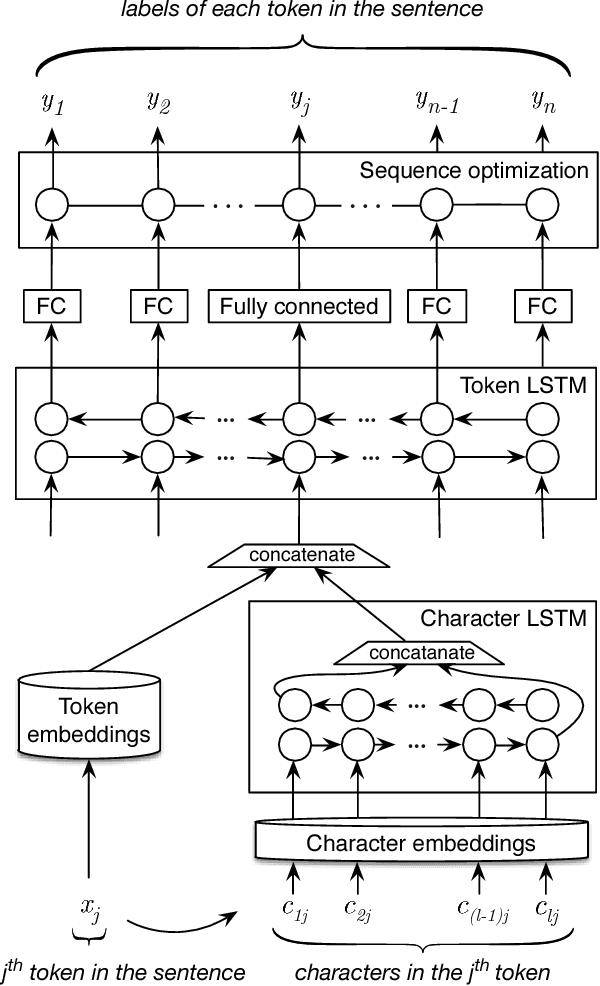
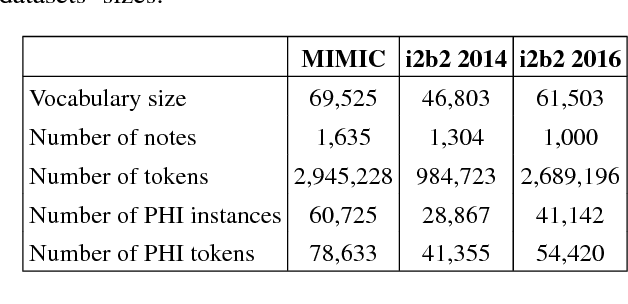
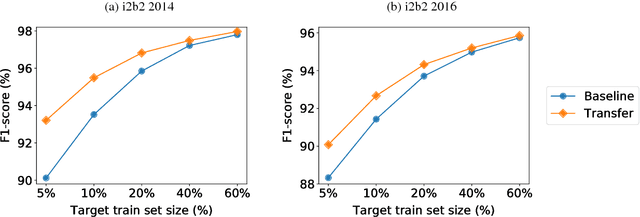
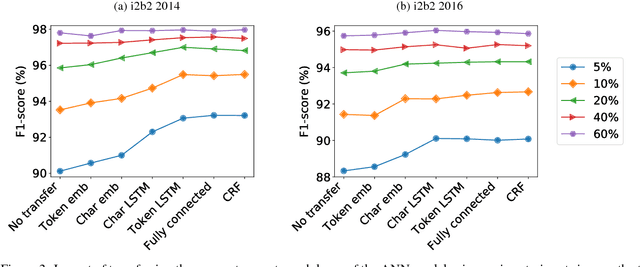
Abstract:Recent approaches based on artificial neural networks (ANNs) have shown promising results for named-entity recognition (NER). In order to achieve high performances, ANNs need to be trained on a large labeled dataset. However, labels might be difficult to obtain for the dataset on which the user wants to perform NER: label scarcity is particularly pronounced for patient note de-identification, which is an instance of NER. In this work, we analyze to what extent transfer learning may address this issue. In particular, we demonstrate that transferring an ANN model trained on a large labeled dataset to another dataset with a limited number of labels improves upon the state-of-the-art results on two different datasets for patient note de-identification.
NeuroNER: an easy-to-use program for named-entity recognition based on neural networks
May 16, 2017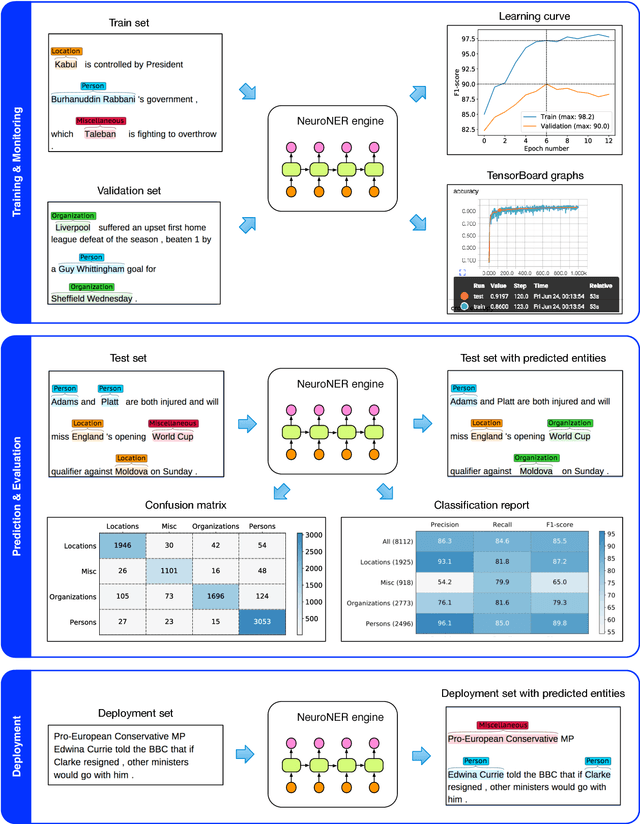
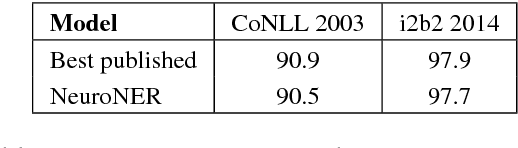
Abstract:Named-entity recognition (NER) aims at identifying entities of interest in a text. Artificial neural networks (ANNs) have recently been shown to outperform existing NER systems. However, ANNs remain challenging to use for non-expert users. In this paper, we present NeuroNER, an easy-to-use named-entity recognition tool based on ANNs. Users can annotate entities using a graphical web-based user interface (BRAT): the annotations are then used to train an ANN, which in turn predict entities' locations and categories in new texts. NeuroNER makes this annotation-training-prediction flow smooth and accessible to anyone.
MIT at SemEval-2017 Task 10: Relation Extraction with Convolutional Neural Networks
Apr 05, 2017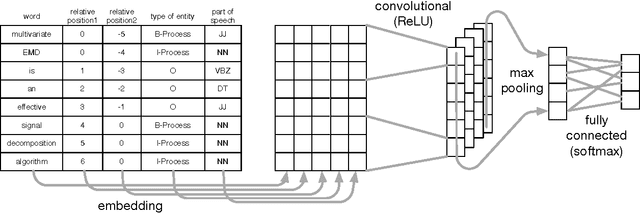
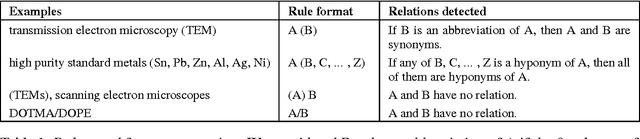
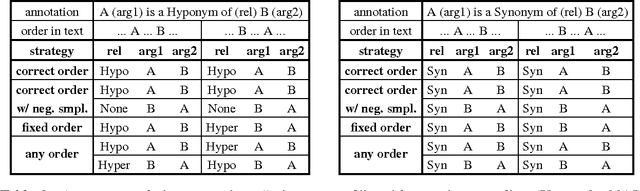
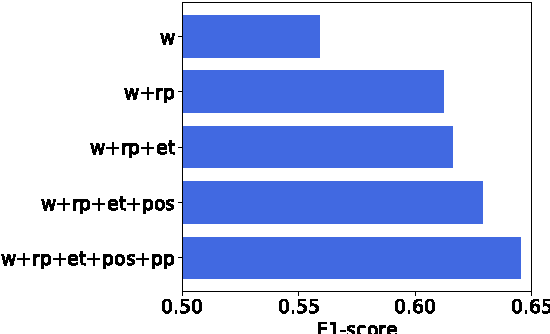
Abstract:Over 50 million scholarly articles have been published: they constitute a unique repository of knowledge. In particular, one may infer from them relations between scientific concepts, such as synonyms and hyponyms. Artificial neural networks have been recently explored for relation extraction. In this work, we continue this line of work and present a system based on a convolutional neural network to extract relations. Our model ranked first in the SemEval-2017 task 10 (ScienceIE) for relation extraction in scientific articles (subtask C).
Neural Networks for Joint Sentence Classification in Medical Paper Abstracts
Dec 15, 2016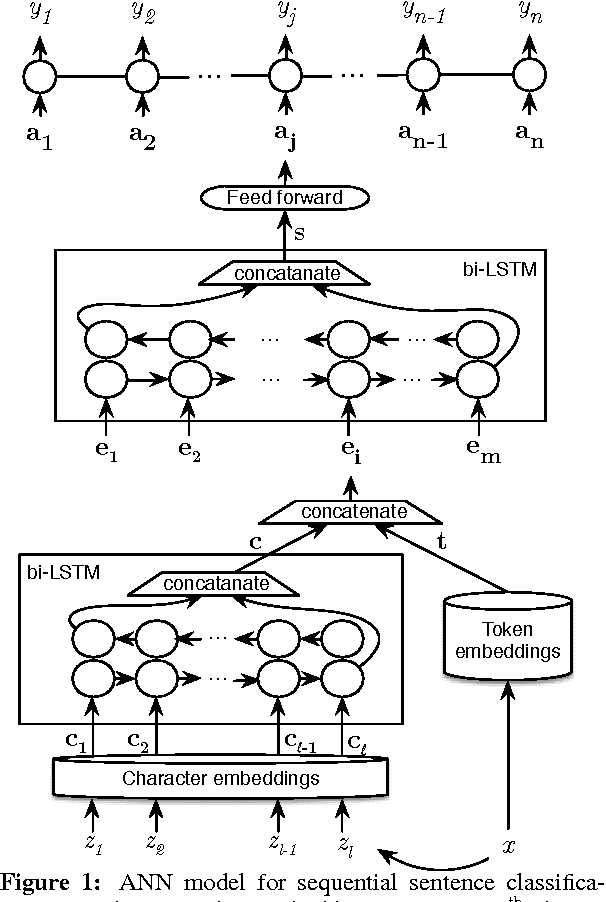
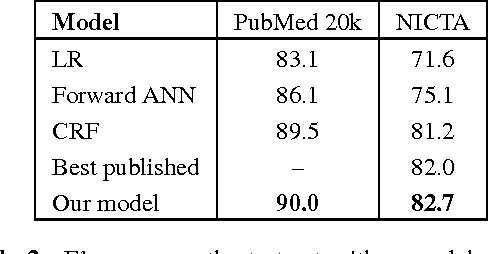
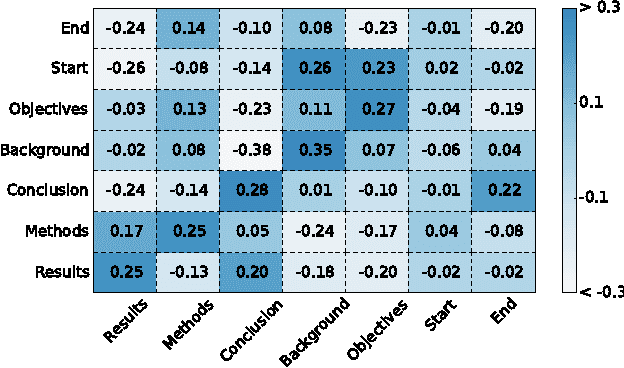
Abstract:Existing models based on artificial neural networks (ANNs) for sentence classification often do not incorporate the context in which sentences appear, and classify sentences individually. However, traditional sentence classification approaches have been shown to greatly benefit from jointly classifying subsequent sentences, such as with conditional random fields. In this work, we present an ANN architecture that combines the effectiveness of typical ANN models to classify sentences in isolation, with the strength of structured prediction. Our model achieves state-of-the-art results on two different datasets for sequential sentence classification in medical abstracts.
Feature-Augmented Neural Networks for Patient Note De-identification
Oct 30, 2016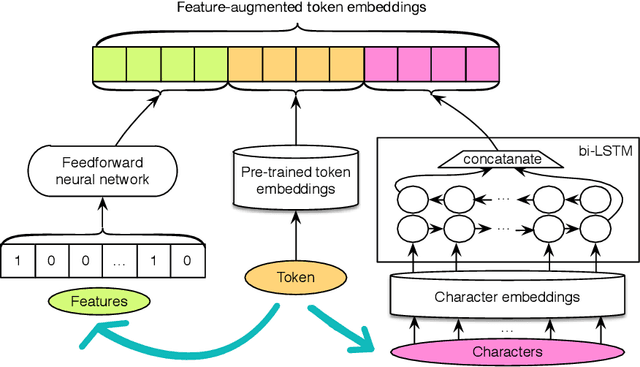

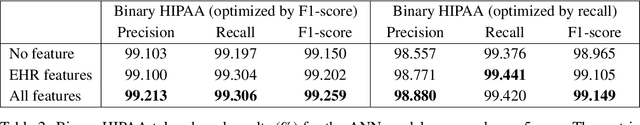
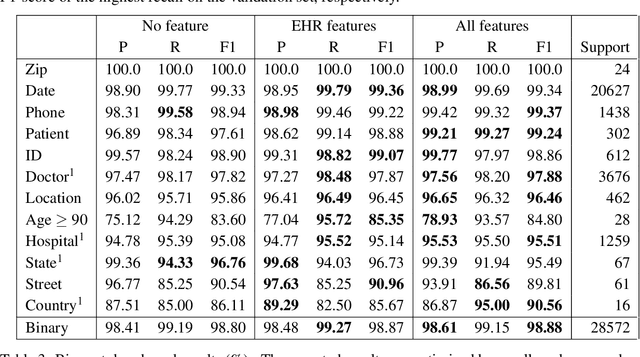
Abstract:Patient notes contain a wealth of information of potentially great interest to medical investigators. However, to protect patients' privacy, Protected Health Information (PHI) must be removed from the patient notes before they can be legally released, a process known as patient note de-identification. The main objective for a de-identification system is to have the highest possible recall. Recently, the first neural-network-based de-identification system has been proposed, yielding state-of-the-art results. Unlike other systems, it does not rely on human-engineered features, which allows it to be quickly deployed, but does not leverage knowledge from human experts or from electronic health records (EHRs). In this work, we explore a method to incorporate human-engineered features as well as features derived from EHRs to a neural-network-based de-identification system. Our results show that the addition of features, especially the EHR-derived features, further improves the state-of-the-art in patient note de-identification, including for some of the most sensitive PHI types such as patient names. Since in a real-life setting patient notes typically come with EHRs, we recommend developers of de-identification systems to leverage the information EHRs contain.
Optimizing Neural Network Hyperparameters with Gaussian Processes for Dialog Act Classification
Sep 27, 2016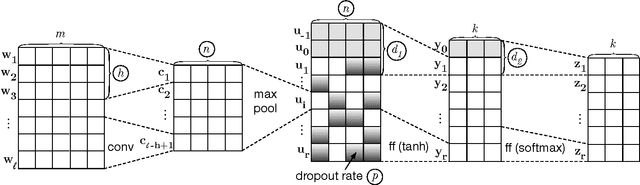
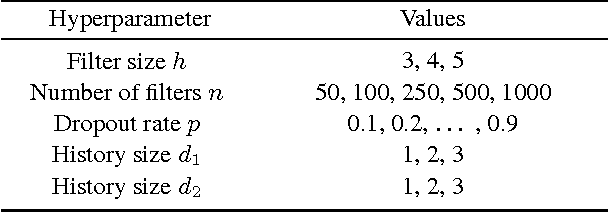
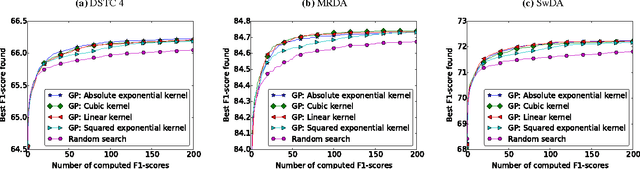
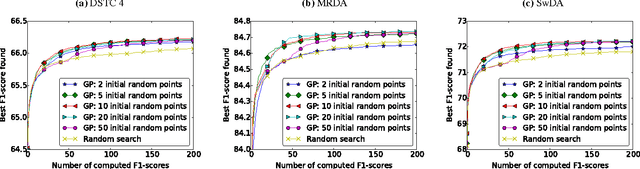
Abstract:Systems based on artificial neural networks (ANNs) have achieved state-of-the-art results in many natural language processing tasks. Although ANNs do not require manually engineered features, ANNs have many hyperparameters to be optimized. The choice of hyperparameters significantly impacts models' performances. However, the ANN hyperparameters are typically chosen by manual, grid, or random search, which either requires expert experiences or is computationally expensive. Recent approaches based on Bayesian optimization using Gaussian processes (GPs) is a more systematic way to automatically pinpoint optimal or near-optimal machine learning hyperparameters. Using a previously published ANN model yielding state-of-the-art results for dialog act classification, we demonstrate that optimizing hyperparameters using GP further improves the results, and reduces the computational time by a factor of 4 compared to a random search. Therefore it is a useful technique for tuning ANN models to yield the best performances for natural language processing tasks.
De-identification of Patient Notes with Recurrent Neural Networks
Jun 10, 2016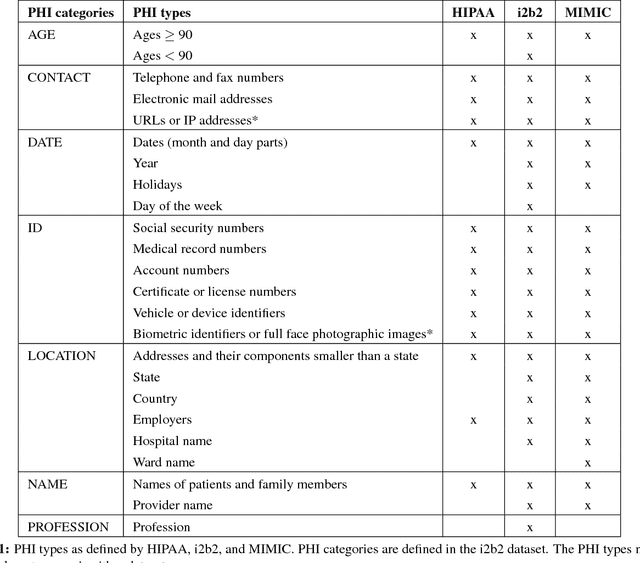
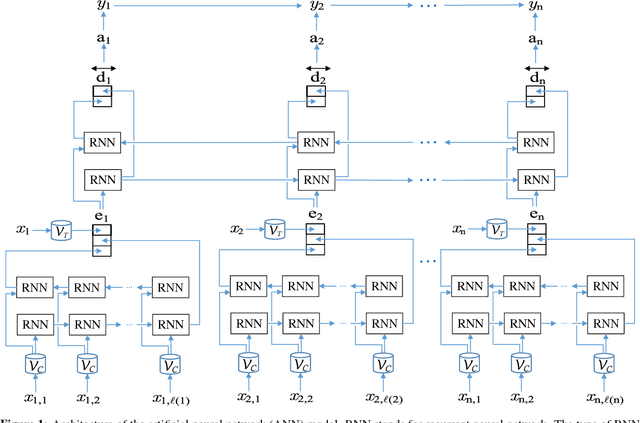
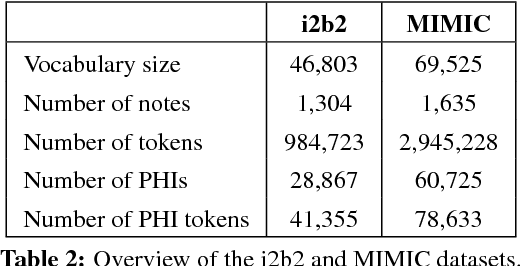
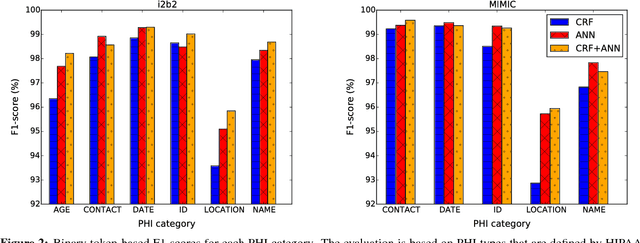
Abstract:Objective: Patient notes in electronic health records (EHRs) may contain critical information for medical investigations. However, the vast majority of medical investigators can only access de-identified notes, in order to protect the confidentiality of patients. In the United States, the Health Insurance Portability and Accountability Act (HIPAA) defines 18 types of protected health information (PHI) that needs to be removed to de-identify patient notes. Manual de-identification is impractical given the size of EHR databases, the limited number of researchers with access to the non-de-identified notes, and the frequent mistakes of human annotators. A reliable automated de-identification system would consequently be of high value. Materials and Methods: We introduce the first de-identification system based on artificial neural networks (ANNs), which requires no handcrafted features or rules, unlike existing systems. We compare the performance of the system with state-of-the-art systems on two datasets: the i2b2 2014 de-identification challenge dataset, which is the largest publicly available de-identification dataset, and the MIMIC de-identification dataset, which we assembled and is twice as large as the i2b2 2014 dataset. Results: Our ANN model outperforms the state-of-the-art systems. It yields an F1-score of 97.85 on the i2b2 2014 dataset, with a recall 97.38 and a precision of 97.32, and an F1-score of 99.23 on the MIMIC de-identification dataset, with a recall 99.25 and a precision of 99.06. Conclusion: Our findings support the use of ANNs for de-identification of patient notes, as they show better performance than previously published systems while requiring no feature engineering.
Robust Dialog State Tracking for Large Ontologies
May 07, 2016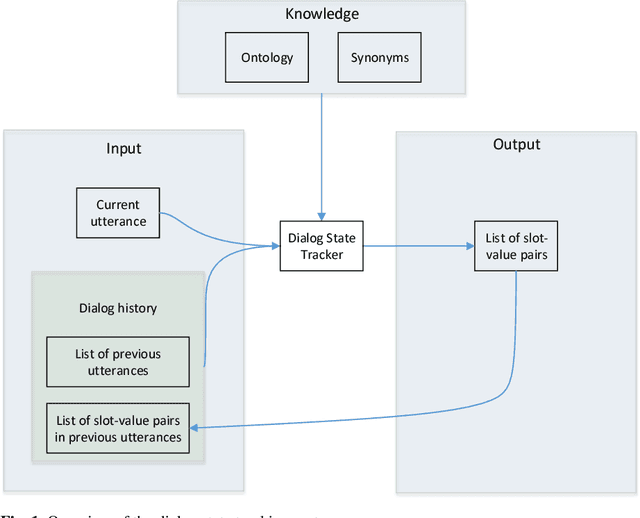
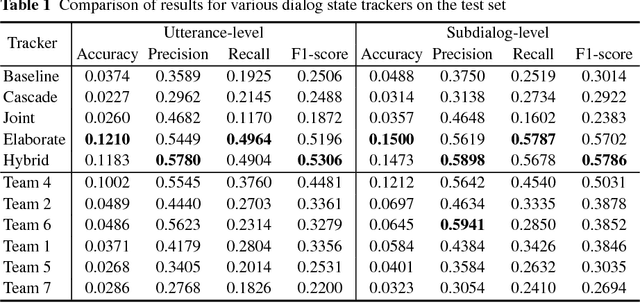
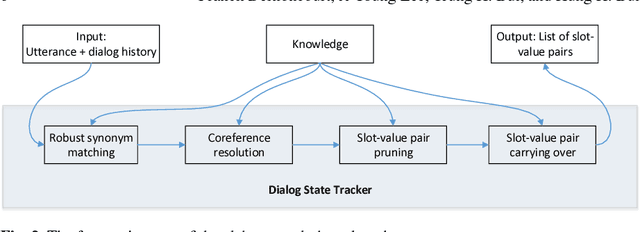
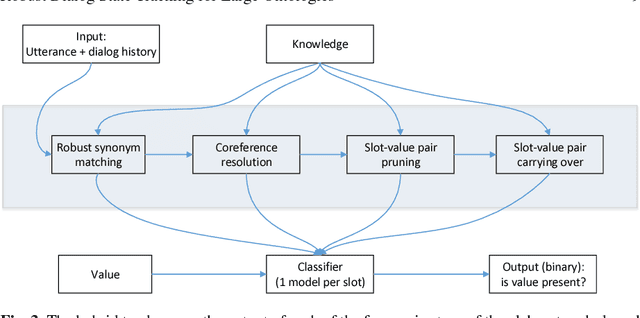
Abstract:The Dialog State Tracking Challenge 4 (DSTC 4) differentiates itself from the previous three editions as follows: the number of slot-value pairs present in the ontology is much larger, no spoken language understanding output is given, and utterances are labeled at the subdialog level. This paper describes a novel dialog state tracking method designed to work robustly under these conditions, using elaborate string matching, coreference resolution tailored for dialogs and a few other improvements. The method can correctly identify many values that are not explicitly present in the utterance. On the final evaluation, our method came in first among 7 competing teams and 24 entries. The F1-score achieved by our method was 9 and 7 percentage points higher than that of the runner-up for the utterance-level evaluation and for the subdialog-level evaluation, respectively.
Adobe-MIT submission to the DSTC 4 Spoken Language Understanding pilot task
May 07, 2016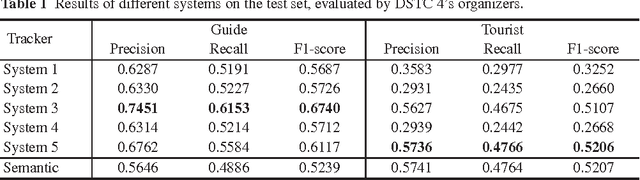
Abstract:The Dialog State Tracking Challenge 4 (DSTC 4) proposes several pilot tasks. In this paper, we focus on the spoken language understanding pilot task, which consists of tagging a given utterance with speech acts and semantic slots. We compare different classifiers: the best system obtains 0.52 and 0.67 F1-scores on the test set for speech act recognition for the tourist and the guide respectively, and 0.52 F1-score for semantic tagging for both the guide and the tourist.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge