James Kapaldo
Seed-Point Based Geometric Partitioning of Nuclei Clumps
Apr 12, 2018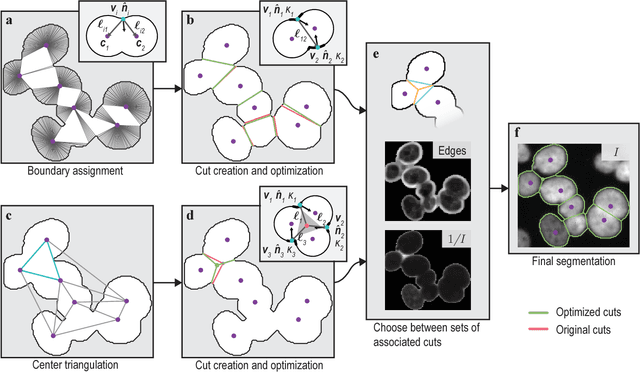
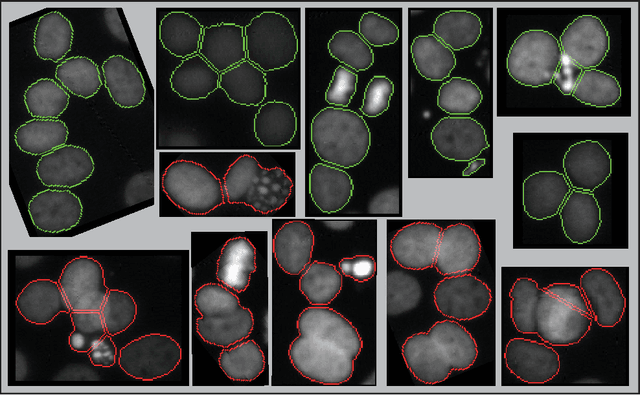
Abstract:When applying automatic analysis of fluorescence or histopathological images of cells, it is necessary to partition, or de-clump, partially overlapping cell nuclei. In this work, I describe a method of partitioning partially overlapping cell nuclei using a seed-point based geometric partitioning. The geometric partitioning creates two different types of cuts, cuts between two boundary vertices and cuts between one boundary vertex and a new vertex introduced to the boundary interior. The cuts are then ranked according to a scoring metric, and the highest scoring cuts are used. This method was tested on a set of 2420 clumps of nuclei and was found to produced better results than current popular analysis software.
Seed-Point Detection of Clumped Convex Objects by Short-Range Attractive Long-Range Repulsive Particle Clustering
Apr 11, 2018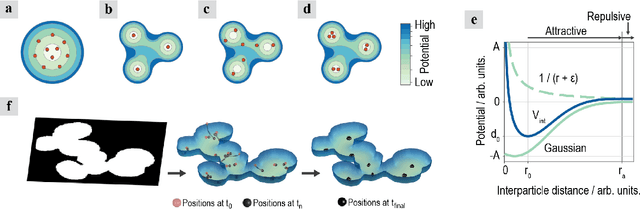

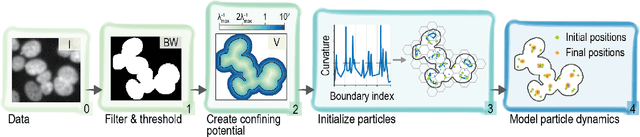

Abstract:Locating the center of convex objects is important in both image processing and unsupervised machine learning/data clustering fields. The automated analysis of biological images uses both of these fields for locating cell nuclei and for discovering new biological effects or cell phenotypes. In this work, we develop a novel clustering method for locating the centers of overlapping convex objects by modeling particles that interact by a short-range attractive and long-range repulsive potential and are confined to a potential well created from the data. We apply this method to locating the centers of clumped nuclei in cultured cells, where we show that it results in a significant improvement over existing methods (8.2% in F$_1$ score); and we apply it to unsupervised learning on a difficult data set that has rare classes without local density maxima, and show it is able to well locate cluster centers when other clustering techniques fail.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge