Jaakko Peltonen
Forecasting Emergency Department Crowding with Advanced Machine Learning Models and Multivariable Input
Aug 31, 2023Abstract:Emergency department (ED) crowding is a significant threat to patient safety and it has been repeatedly associated with increased mortality. Forecasting future service demand has the potential patient outcomes. Despite active research on the subject, several gaps remain: 1) proposed forecasting models have become outdated due to quick influx of advanced machine learning models (ML), 2) amount of multivariable input data has been limited and 3) discrete performance metrics have been rarely reported. In this study, we document the performance of a set of advanced ML models in forecasting ED occupancy 24 hours ahead. We use electronic health record data from a large, combined ED with an extensive set of explanatory variables, including the availability of beds in catchment area hospitals, traffic data from local observation stations, weather variables, etc. We show that N-BEATS and LightGBM outpeform benchmarks with 11 % and 9 % respective improvements and that DeepAR predicts next day crowding with an AUC of 0.76 (95 % CI 0.69-0.84). To the best of our knowledge, this is the first study to document the superiority of LightGBM and N-BEATS over statistical benchmarks in the context of ED forecasting.
Scalable Probabilistic Matrix Factorization with Graph-Based Priors
Sep 11, 2019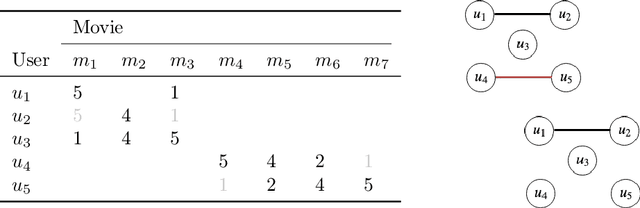
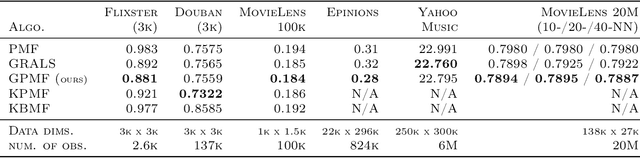
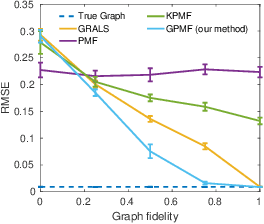
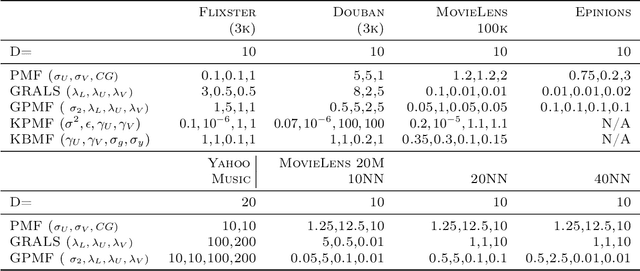
Abstract:In matrix factorization, available graph side-information may not be well suited for the matrix completion problem, having edges that disagree with the latent-feature relations learnt from the incomplete data matrix. We show that removing these $\textit{contested}$ edges improves prediction accuracy and scalability. We identify the contested edges through a highly-efficient graphical lasso approximation. The identification and removal of contested edges adds no computational complexity to state-of-the-art graph-regularized matrix factorization, remaining linear with respect to the number of non-zeros. Computational load even decreases proportional to the number of edges removed. Formulating a probabilistic generative model and using expectation maximization to extend graph-regularised alternating least squares (GRALS) guarantees convergence. Rich simulated experiments illustrate the desired properties of the resulting algorithm. On real data experiments we demonstrate improved prediction accuracy with fewer graph edges (empirical evidence that graph side-information is often inaccurate). A 300 thousand dimensional graph with three million edges (Yahoo music side-information) can be analyzed in under ten minutes on a standard laptop computer demonstrating the efficiency of our graph update.
Peacock Bundles: Bundle Coloring for Graphs with Globality-Locality Trade-off
Sep 02, 2016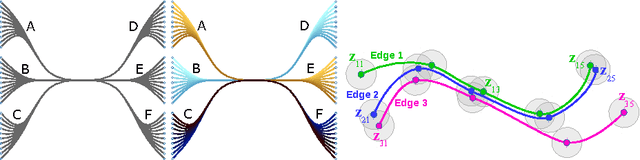
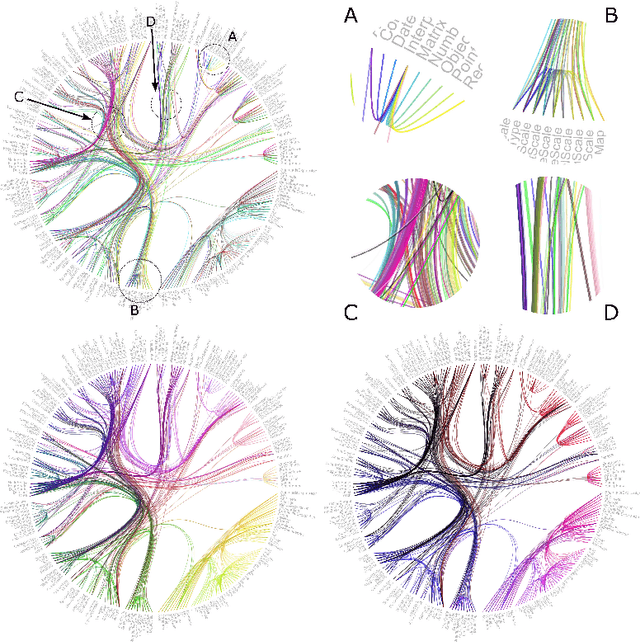
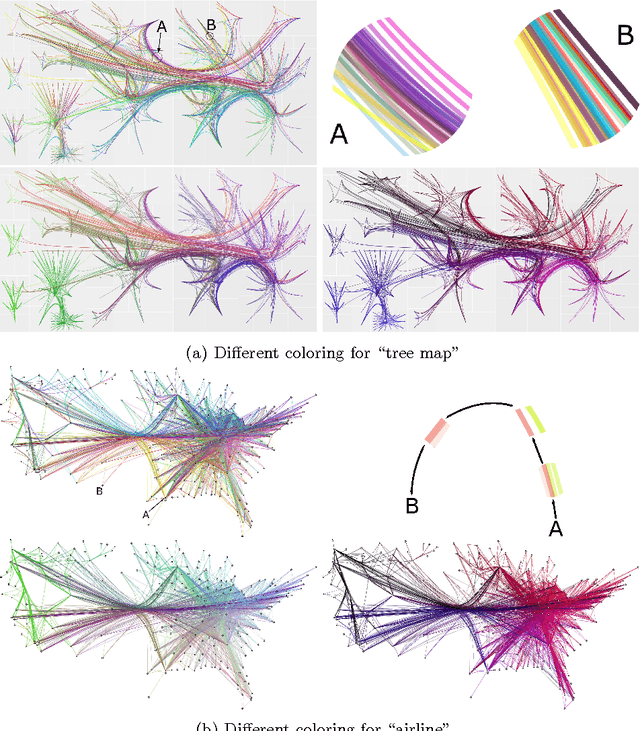

Abstract:Bundling of graph edges (node-to-node connections) is a common technique to enhance visibility of overall trends in the edge structure of a large graph layout, and a large variety of bundling algorithms have been proposed. However, with strong bundling, it becomes hard to identify origins and destinations of individual edges. We propose a solution: we optimize edge coloring to differentiate bundled edges. We quantify strength of bundling in a flexible pairwise fashion between edges, and among bundled edges, we quantify how dissimilar their colors should be by dissimilarity of their origins and destinations. We solve the resulting nonlinear optimization, which is also interpretable as a novel dimensionality reduction task. In large graphs the necessary compromise is whether to differentiate colors sharply between locally occurring strongly bundled edges ("local bundles"), or also between the weakly bundled edges occurring globally over the graph ("global bundles"); we allow a user-set global-local tradeoff. We call the technique "peacock bundles". Experiments show the coloring clearly enhances comprehensibility of graph layouts with edge bundling.
An Information Retrieval Approach to Finding Dependent Subspaces of Multiple Views
Jan 07, 2016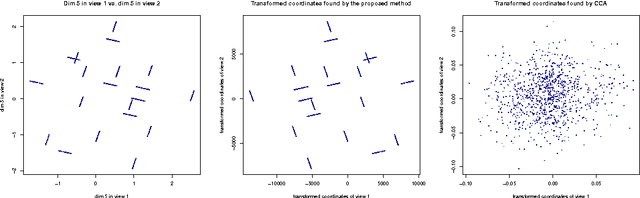
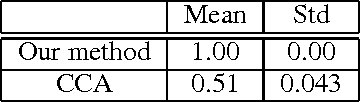
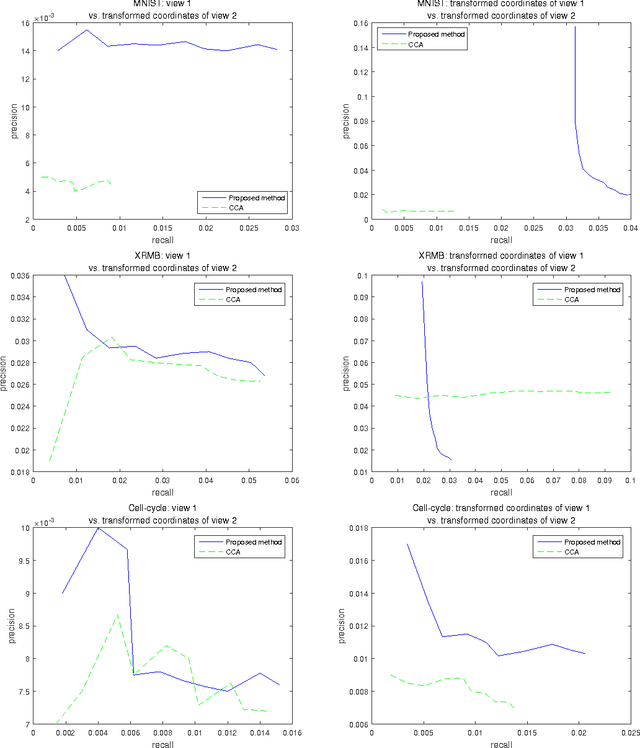
Abstract:Finding relationships between multiple views of data is essential both for exploratory analysis and as pre-processing for predictive tasks. A prominent approach is to apply variants of Canonical Correlation Analysis (CCA), a classical method seeking correlated components between views. The basic CCA is restricted to maximizing a simple dependency criterion, correlation, measured directly between data coordinates. We introduce a new method that finds dependent subspaces of views directly optimized for the data analysis task of \textit{neighbor retrieval between multiple views}. We optimize mappings for each view such as linear transformations to maximize cross-view similarity between neighborhoods of data samples. The criterion arises directly from the well-defined retrieval task, detects nonlinear and local similarities, is able to measure dependency of data relationships rather than only individual data coordinates, and is related to well understood measures of information retrieval quality. In experiments we show the proposed method outperforms alternatives in preserving cross-view neighborhood similarities, and yields insights into local dependencies between multiple views.
Toward computational cumulative biology by combining models of biological datasets
Apr 01, 2014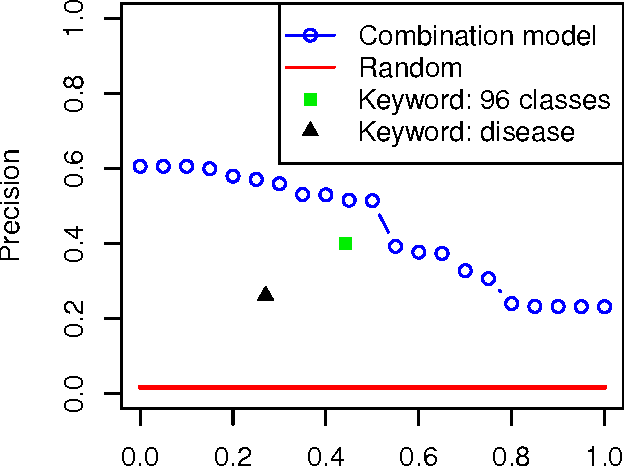
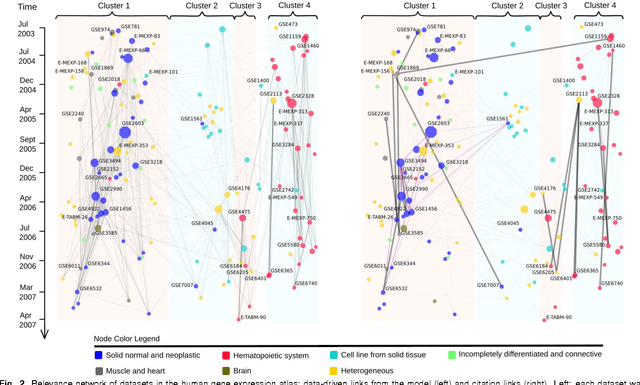
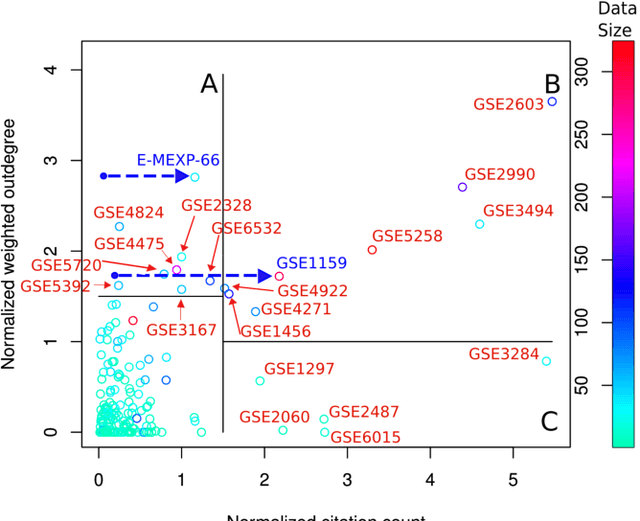
Abstract:A main challenge of data-driven sciences is how to make maximal use of the progressively expanding databases of experimental datasets in order to keep research cumulative. We introduce the idea of a modeling-based dataset retrieval engine designed for relating a researcher's experimental dataset to earlier work in the field. The search is (i) data-driven to enable new findings, going beyond the state of the art of keyword searches in annotations, (ii) modeling-driven, to both include biological knowledge and insights learned from data, and (iii) scalable, as it is accomplished without building one unified grand model of all data. Assuming each dataset has been modeled beforehand, by the researchers or by database managers, we apply a rapidly computable and optimizable combination model to decompose a new dataset into contributions from earlier relevant models. By using the data-driven decomposition we identify a network of interrelated datasets from a large annotated human gene expression atlas. While tissue type and disease were major driving forces for determining relevant datasets, the found relationships were richer and the model-based search was more accurate than keyword search; it moreover recovered biologically meaningful relationships that are not straightforwardly visible from annotations, for instance, between cells in different developmental stages such as thymocytes and T-cells. Data-driven links and citations matched to a large extent; the data-driven links even uncovered corrections to the publication data, as two of the most linked datasets were not highly cited and turned out to have wrong publication entries in the database.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge