Jürgen Weese
3D medical image segmentation with labeled and unlabeled data using autoencoders at the example of liver segmentation in CT images
Mar 17, 2020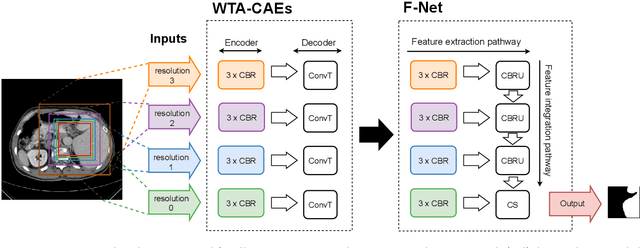
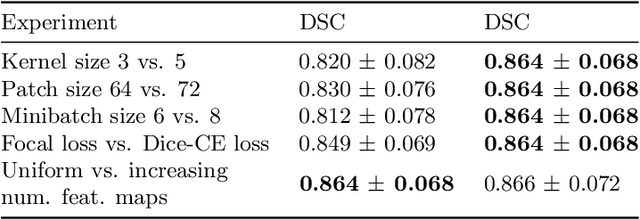

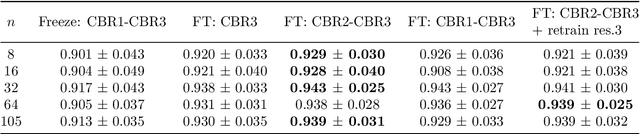
Abstract:Automatic segmentation of anatomical structures with convolutional neural networks (CNNs) constitutes a large portion of research in medical image analysis. The majority of CNN-based methods rely on an abundance of labeled data for proper training. Labeled medical data is often scarce, but unlabeled data is more widely available. This necessitates approaches that go beyond traditional supervised learning and leverage unlabeled data for segmentation tasks. This work investigates the potential of autoencoder-extracted features to improve segmentation with a CNN. Two strategies were considered. First, transfer learning where pretrained autoencoder features were used as initialization for the convolutional layers in the segmentation network. Second, multi-task learning where the tasks of segmentation and feature extraction, by means of input reconstruction, were learned and optimized simultaneously. A convolutional autoencoder was used to extract features from unlabeled data and a multi-scale, fully convolutional CNN was used to perform the target task of 3D liver segmentation in CT images. For both strategies, experiments were conducted with varying amounts of labeled and unlabeled training data. The proposed learning strategies improved results in $75\%$ of the experiments compared to training from scratch and increased the dice score by up to $0.040$ and $0.024$ for a ratio of unlabeled to labeled training data of about $32 : 1$ and $12.5 : 1$, respectively. The results indicate that both training strategies are more effective with a large ratio of unlabeled to labeled training data.
Iterative Segmentation from Limited Training Data: Applications to Congenital Heart Disease
Sep 11, 2018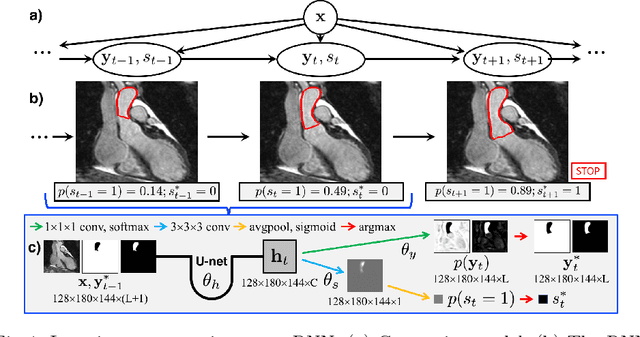


Abstract:We propose a new iterative segmentation model which can be accurately learned from a small dataset. A common approach is to train a model to directly segment an image, requiring a large collection of manually annotated images to capture the anatomical variability in a cohort. In contrast, we develop a segmentation model that recursively evolves a segmentation in several steps, and implement it as a recurrent neural network. We learn model parameters by optimizing the interme- diate steps of the evolution in addition to the final segmentation. To this end, we train our segmentation propagation model by presenting incom- plete and/or inaccurate input segmentations paired with a recommended next step. Our work aims to alleviate challenges in segmenting heart structures from cardiac MRI for patients with congenital heart disease (CHD), which encompasses a range of morphological deformations and topological changes. We demonstrate the advantages of this approach on a dataset of 20 images from CHD patients, learning a model that accurately segments individual heart chambers and great vessels. Com- pared to direct segmentation, the iterative method yields more accurate segmentation for patients with the most severe CHD malformations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge