Jürgen Wallner
Exploit fully automatic low-level segmented PET data for training high-level deep learning algorithms for the corresponding CT data
Mar 07, 2019



Abstract:We present an approach for fully automatic urinary bladder segmentation in CT images with artificial neural networks in this study. Automatic medical image analysis has become an invaluable tool in the different treatment stages of diseases. Especially medical image segmentation plays a vital role, since segmentation is often the initial step in an image analysis pipeline. Since deep neural networks have made a large impact on the field of image processing in the past years, we use two different deep learning architectures to segment the urinary bladder. Both of these architectures are based on pre-trained classification networks that are adapted to perform semantic segmentation. Since deep neural networks require a large amount of training data, specifically images and corresponding ground truth labels, we furthermore propose a method to generate such a suitable training data set from Positron Emission Tomography/Computed Tomography image data. This is done by applying thresholding to the Positron Emission Tomography data for obtaining a ground truth and by utilizing data augmentation to enlarge the dataset. In this study, we discuss the influence of data augmentation on the segmentation results, and compare and evaluate the proposed architectures in terms of qualitative and quantitative segmentation performance. The results presented in this study allow concluding that deep neural networks can be considered a promising approach to segment the urinary bladder in CT images.
* 20 pages
Computed tomography data collection of the complete human mandible and valid clinical ground truth models
Feb 14, 2019



Abstract:Image-based algorithmic software segmentation is an increasingly important topic in many medical fields. Algorithmic segmentation is used for medical three-dimensional visualization, diagnosis or treatment support, especially in complex medical cases. However, accessible medical databases are limited, and valid medical ground truth databases for the evaluation of algorithms are rare and usually comprise only a few images. Inaccuracy or invalidity of medical ground truth data and image-based artefacts also limit the creation of such databases, which is especially relevant for CT data sets of the maxillomandibular complex. This contribution provides a unique and accessible data set of the complete mandible, including 20 valid ground truth segmentation models originating from 10 CT scans from clinical practice without artefacts or faulty slices. From each CT scan, two 3D ground truth models were created by clinical experts through independent manual slice-by-slice segmentation, and the models were statistically compared to prove their validity. These data could be used to conduct serial image studies of the human mandible, evaluating segmentation algorithms and developing adequate image tools.
* 14 pages, 8 Figures, 4 Tables, 55 References
Clinical evaluation of semi-automatic opensource algorithmic software segmentation of the mandibular bone: Practical feasibility and assessment of a new course of action
May 11, 2018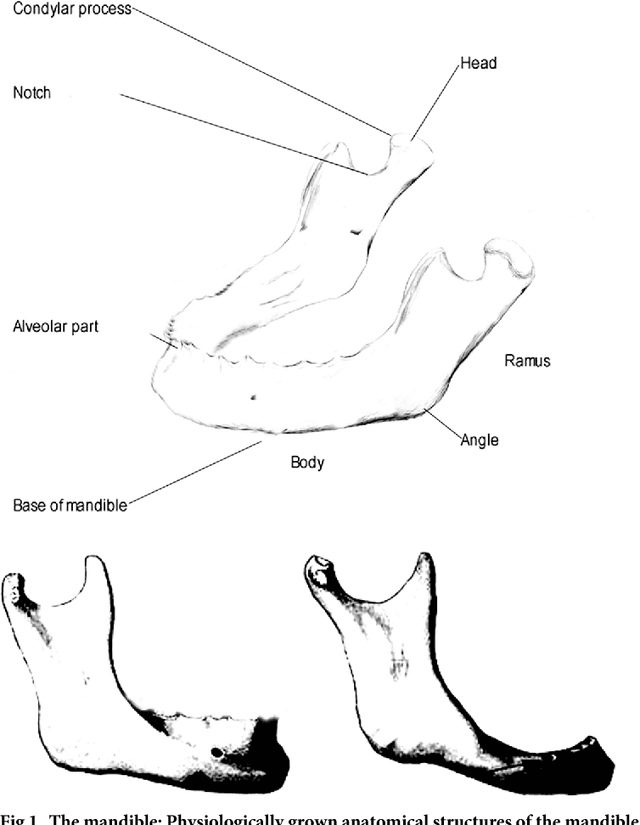
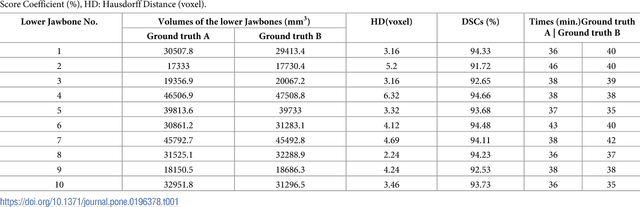
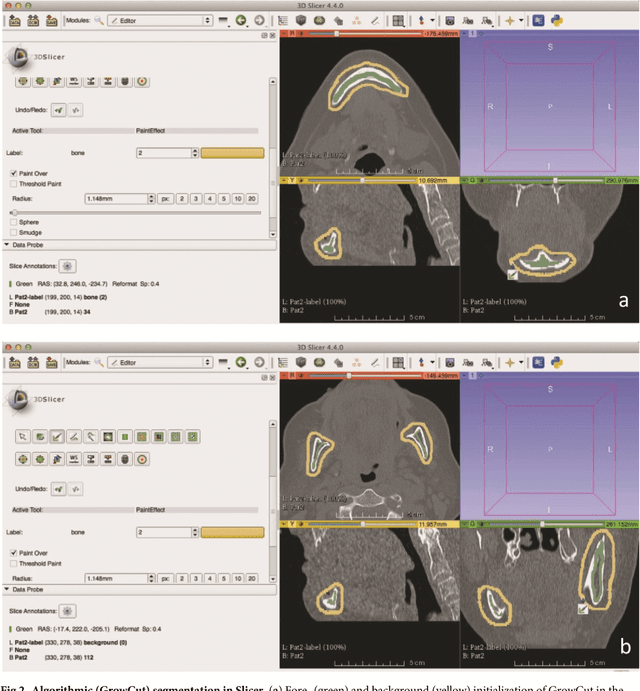
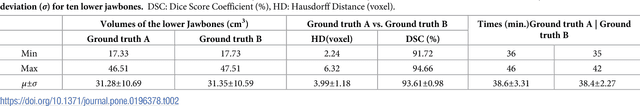
Abstract:Computer assisted technologies based on algorithmic software segmentation are an increasing topic of interest in complex surgical cases. However - due to functional instability, time consuming software processes, personnel resources or licensed-based financial costs many segmentation processes are often outsourced from clinical centers to third parties and the industry. Therefore, the aim of this trial was to assess the practical feasibility of an easy available, functional stable and licensed-free segmentation approach to be used in the clinical practice. In this retrospective, randomized, controlled trail the accuracy and accordance of the open-source based segmentation algorithm GrowCut (GC) was assessed through the comparison to the manually generated ground truth of the same anatomy using 10 CT lower jaw data-sets from the clinical routine. Assessment parameters were the segmentation time, the volume, the voxel number, the Dice Score (DSC) and the Hausdorff distance (HD). Overall segmentation times were about one minute. Mean DSC values of over 85% and HD below 33.5 voxel could be achieved. Statistical differences between the assessment parameters were not significant (p<0.05) and correlation coefficients were close to the value one (r > 0.94). Complete functional stable and time saving segmentations with high accuracy and high positive correlation could be performed by the presented interactive open-source based approach. In the cranio-maxillofacial complex the used method could represent an algorithmic alternative for image-based segmentation in the clinical practice for e.g. surgical treatment planning or visualization of postoperative results and offers several advantages. Systematic comparisons to other segmentation approaches or with a greater data amount are areas of future works.
* 26 pages
Computer-aided position planning of miniplates to treat facial bone defects
Aug 17, 2017



Abstract:In this contribution, a software system for computer-aided position planning of miniplates to treat facial bone defects is proposed. The intra-operatively used bone plates have to be passively adapted on the underlying bone contours for adequate bone fragment stabilization. However, this procedure can lead to frequent intra-operatively performed material readjustments especially in complex surgical cases. Our approach is able to fit a selection of common implant models on the surgeon's desired position in a 3D computer model. This happens with respect to the surrounding anatomical structures, always including the possibility of adjusting both the direction and the position of the used osteosynthesis material. By using the proposed software, surgeons are able to pre-plan the out coming implant in its form and morphology with the aid of a computer-visualized model within a few minutes. Further, the resulting model can be stored in STL file format, the commonly used format for 3D printing. Using this technology, surgeons are able to print the virtual generated implant, or create an individually designed bending tool. This method leads to adapted osteosynthesis materials according to the surrounding anatomy and requires further a minimum amount of money and time.
* 19 pages, 13 Figures, 2 Tables
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge