Isky Gorden
Automatic Delineation of Kidney Region in DCE-MRI
May 26, 2019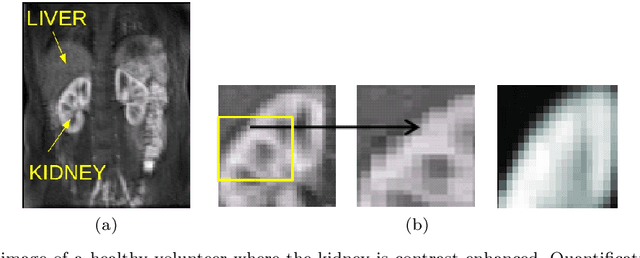
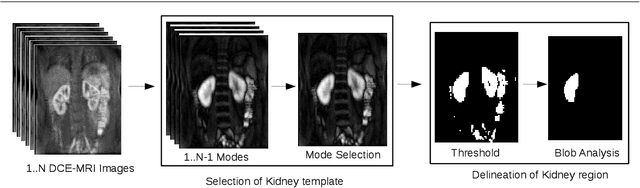
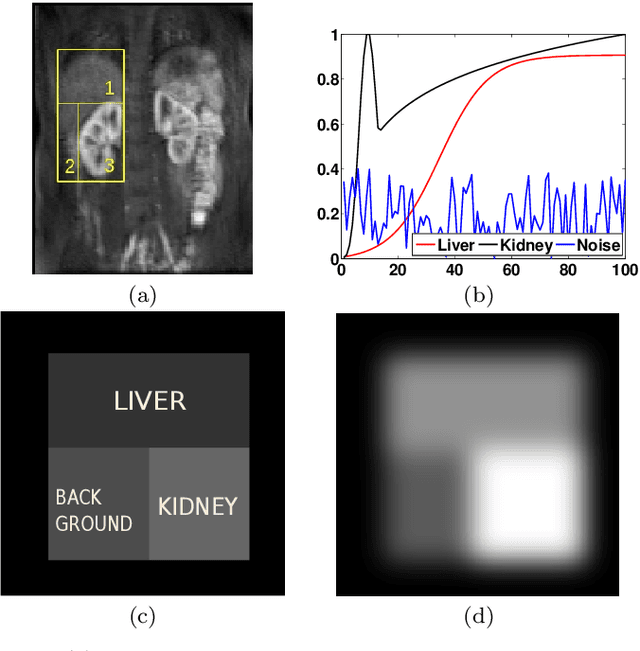
Abstract:Delineation of the kidney region in dynamic contrast-enhanced magnetic resonance Imaging (DCE-MRI) is required during post-acquisition analysis in order to quantify various aspects of renal function, such as filtration and perfusion or blood flow. However, this can be obfuscated by the Partial Volume Effect (PVE), caused due to the mixing of any single voxel with two or more signal intensities from adjacent regions such as liver region and other tissues. To avoid this problem, firstly, a kidney region of interest (ROI) needs to be defined for the analysis. A clinician may choose to select a region avoiding edges where PV mixing is likely to be significant. However, this approach is time-consuming and labour intensive. To address this issue, we present Dynamic Mode Decomposition (DMD) coupled with thresholding and blob analysis as a framework for automatic delineation of the kidney region. This method is first validated on synthetically generated data with ground-truth available and then applied to ten healthy volunteers' kidney DCE-MRI datasets. We found that the result obtained from our proposed framework is comparable to that of a human expert. For example, while our result gives an average Root Mean Square Error (RMSE) of 0.0097, the baseline achieves an average RMSE of 0.1196 across the 10 datasets. As a result, we conclude automatic modelling via DMD framework is a promising approach.
Functional Segmentation through Dynamic Mode Decomposition: Automatic Quantification of Kidney Function in DCE-MRI Images
May 24, 2019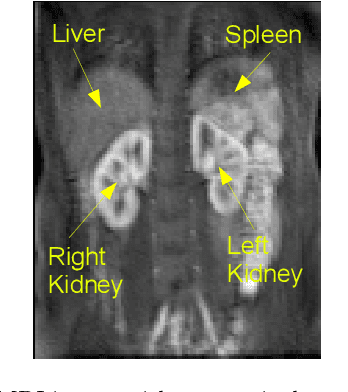
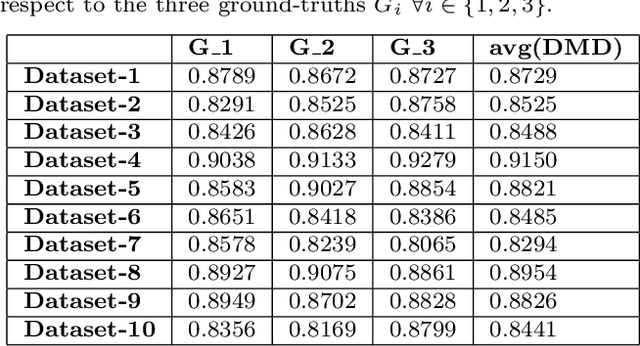
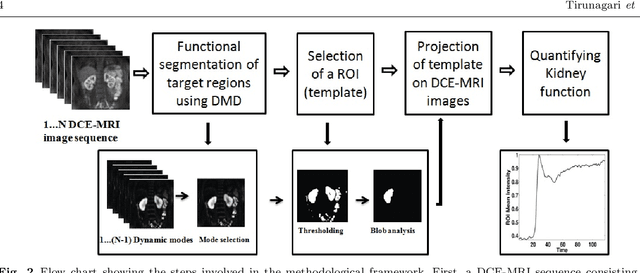
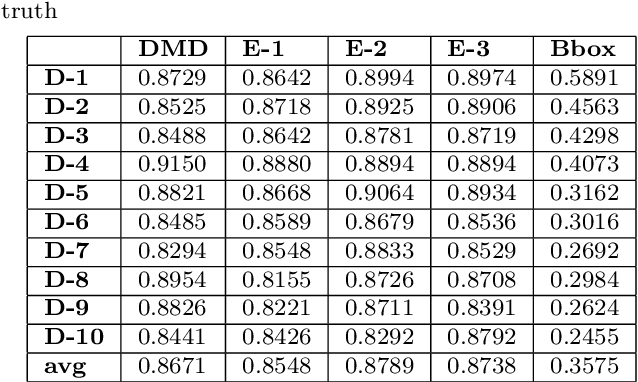
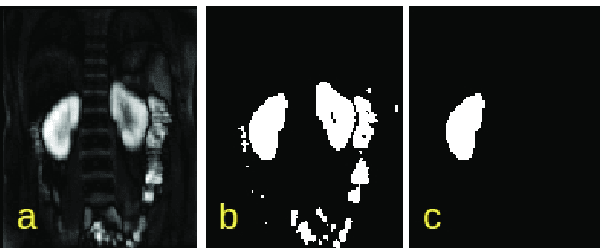
Abstract:Quantification of kidney function in Dynamic Contrast-Enhanced Magnetic Resonance Imaging (DCE-MRI) requires careful segmentation of the renal region of interest (ROI). Traditionally, human experts are required to manually delineate the kidney ROI across multiple images in the dynamic sequence. This approach is costly, time-consuming and labour intensive, and therefore acts to limit patient throughout and acts as one of the factors limiting the wider adoption of DCR-MRI in clinical practice. Therefore, to address this issue, we present the first use of Dynamic Mode Decomposition (DMD) as a basis for automatic segmentation of a dynamic sequence, in this case, kidney ROIs in DCE-MRI. Using DMD coupled combined with thresholding and connected component analysis is first validated on synthetically generated data with known ground-truth, and then applied to ten healthy volunteers' DCE-MRI datasets. We find that the segmentation result obtained from our proposed DMD framework is comparable to that of expert observers and very significantly better than that of an a-priori bounding box segmentation. Our result gives a mean Jaccard coefficient of 0.87, compared to mean scores of 0.85, 0.88 and 0.87 produced from three independent manual annotations. This represents the first use of DMD as a robust automatic data-driven segmentation approach without requiring any human intervention. This is a viable, efficient alternative approach to current manual methods of isolation of kidney function in DCE-MRI.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge