Isamu Isozaki
Towards Automated Penetration Testing: Introducing LLM Benchmark, Analysis, and Improvements
Oct 22, 2024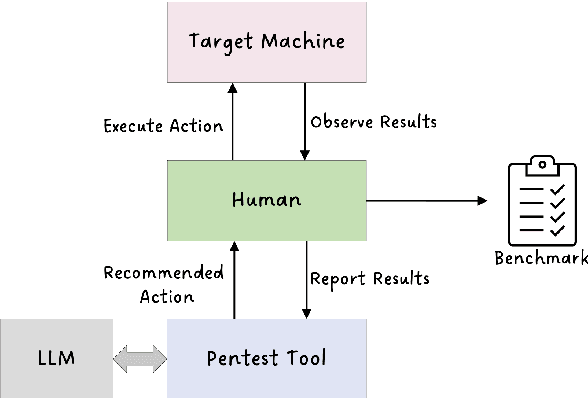
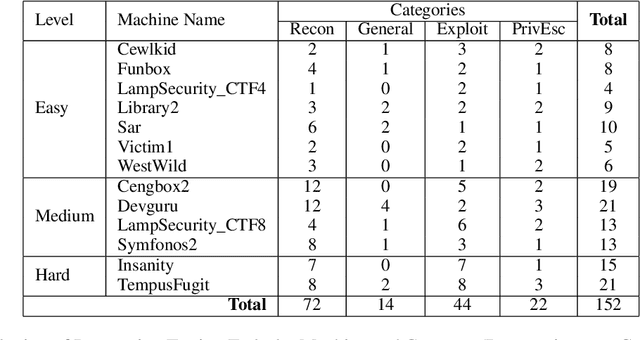
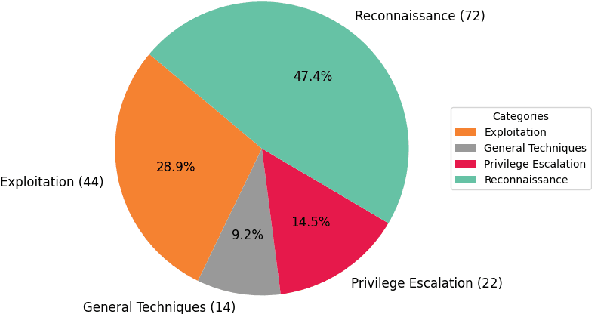
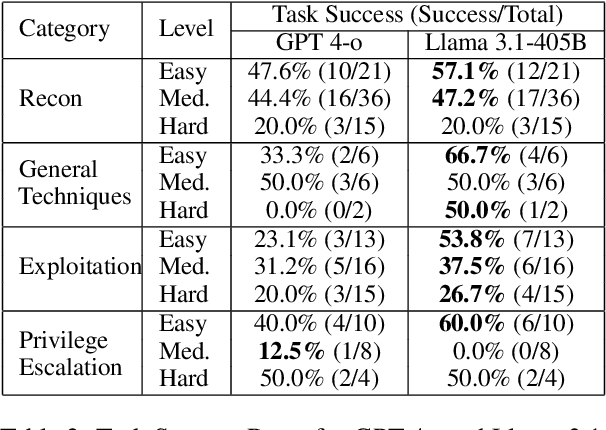
Abstract:Hacking poses a significant threat to cybersecurity, inflicting billions of dollars in damages annually. To mitigate these risks, ethical hacking, or penetration testing, is employed to identify vulnerabilities in systems and networks. Recent advancements in large language models (LLMs) have shown potential across various domains, including cybersecurity. However, there is currently no comprehensive, open, end-to-end automated penetration testing benchmark to drive progress and evaluate the capabilities of these models in security contexts. This paper introduces a novel open benchmark for LLM-based automated penetration testing, addressing this critical gap. We first evaluate the performance of LLMs, including GPT-4o and Llama 3.1-405B, using the state-of-the-art PentestGPT tool. Our findings reveal that while Llama 3.1 demonstrates an edge over GPT-4o, both models currently fall short of performing fully automated, end-to-end penetration testing. Next, we advance the state-of-the-art and present ablation studies that provide insights into improving the PentestGPT tool. Our research illuminates the challenges LLMs face in each aspect of Pentesting, e.g. enumeration, exploitation, and privilege escalation. This work contributes to the growing body of knowledge on AI-assisted cybersecurity and lays the foundation for future research in automated penetration testing using large language models.
Functional Protein Structure Annotation Using a Deep Convolutional Generative Adversarial Network
Apr 18, 2021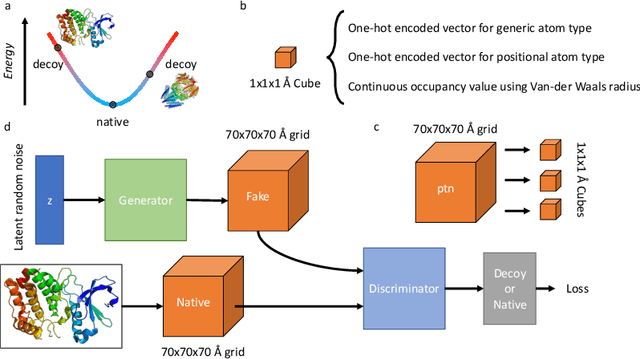
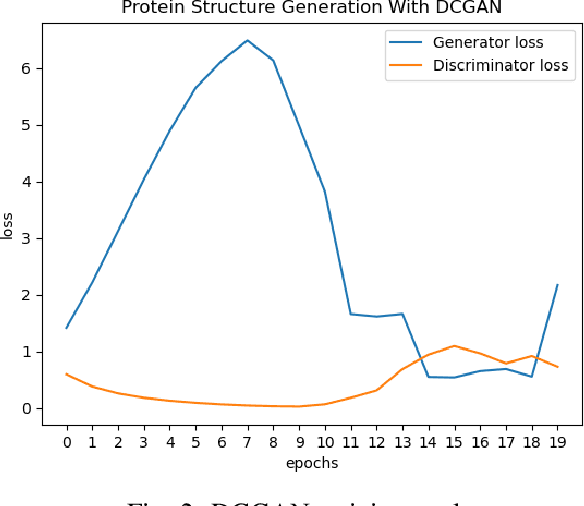
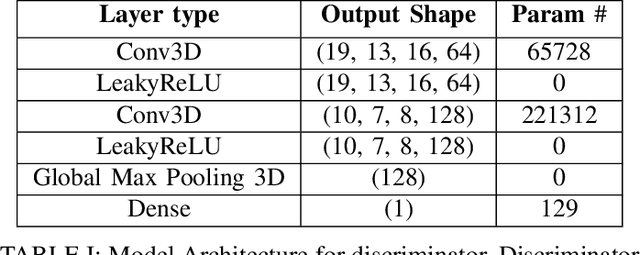
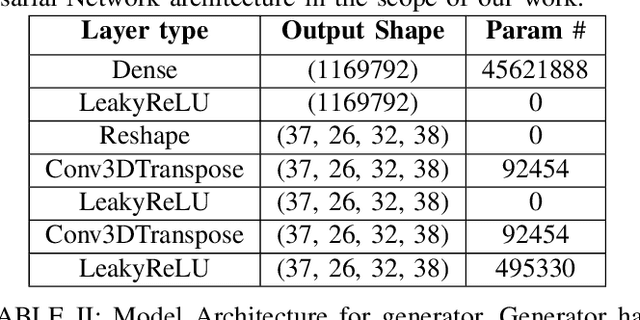
Abstract:Identifying novel functional protein structures is at the heart of molecular engineering and molecular biology, requiring an often computationally exhaustive search. We introduce the use of a Deep Convolutional Generative Adversarial Network (DCGAN) to classify protein structures based on their functionality by encoding each sample in a grid object structure using three features in each object: the generic atom type, the position atom type, and its occupancy relative to a given atom. We train DCGAN on 3-dimensional (3D) decoy and native protein structures in order to generate and discriminate 3D protein structures. At the end of our training, loss converges to a local minimum and our DCGAN can annotate functional proteins robustly against adversarial protein samples. In the future we hope to extend the novel structures we found from the generator in our DCGAN with more samples to explore more granular functionality with varying functions. We hope that our effort will advance the field of protein structure prediction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge