Ipek Oruc
SynapsNet: Enhancing Neuronal Population Dynamics Modeling via Learning Functional Connectivity
Nov 12, 2024



Abstract:The availability of large-scale neuronal population datasets necessitates new methods to model population dynamics and extract interpretable, scientifically translatable insights. Existing deep learning methods often overlook the biological mechanisms underlying population activity and thus exhibit suboptimal performance with neuronal data and provide little to no interpretable information about neurons and their interactions. In response, we introduce SynapsNet, a novel deep-learning framework that effectively models population dynamics and functional interactions between neurons. Within this biologically realistic framework, each neuron, characterized by a latent embedding, sends and receives currents through directed connections. A shared decoder uses the input current, previous neuronal activity, neuron embedding, and behavioral data to predict the population activity in the next time step. Unlike common sequential models that treat population activity as a multichannel time series, SynapsNet applies its decoder to each neuron (channel) individually, with the learnable functional connectivity serving as the sole pathway for information flow between neurons. Our experiments, conducted on mouse cortical activity from publicly available datasets and recorded using the two most common population recording modalities (Ca imaging and Neuropixels) across three distinct tasks, demonstrate that SynapsNet consistently outperforms existing models in forecasting population activity. Additionally, our experiments on both real and synthetic data showed that SynapsNet accurately learns functional connectivity that reveals predictive interactions between neurons.
Artificial intelligence as a gateway to scientific discovery: Uncovering features in retinal fundus images
Jan 17, 2023



Abstract:Purpose: Convolutional neural networks can be trained to detect various conditions or patient traits based on retinal fundus photographs, some of which, such as the patient sex, are invisible to the expert human eye. Here we propose a methodology for explainable classification of fundus images to uncover the mechanism(s) by which CNNs successfully predict the labels. We used patient sex as a case study to validate our proposed methodology. Approach: First, we used a set of 4746 fundus images, including training, validation and test partitions, to fine-tune a pre-trained CNN on the sex classification task. Next, we utilized deep learning explainability tools to hypothesize possible ways sex differences in the retina manifest. We measured numerous retinal properties relevant to our hypotheses through image segmentation to identify those significantly different between males and females. To tackle the multiple comparisons problem, we shortlisted the parameters by testing them on a set of 100 fundus images distinct from the images used for fine-tuning. Finally, we used an additional 400 images, not included in any previous set, to reveal significant sex differences in the retina. Results: We observed that the peripapillary area is darker in males compared to females ($p=.023, d=.243$). We also observed that males have richer retinal vasculature networks by showing a higher number of branches ($p=.016, d=.272$) and nodes ($p=.014, d=.299$) and a larger total length of branches ($p=.045, d=.206$) in the vessel graph. Also, vessels cover a greater area in the superior temporal quadrant of the retina in males compared to females ($p=0.048, d=.194$). Conclusions: Our methodology reveals retinal features in fundus photographs that allow CNNs to predict traits currently unknown, but meaningful to experts.
Learning from few examples: Classifying sex from retinal images via deep learning
Jul 20, 2022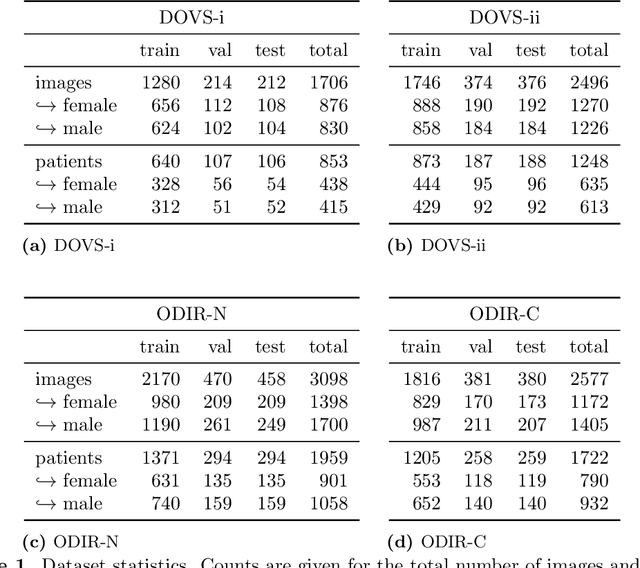

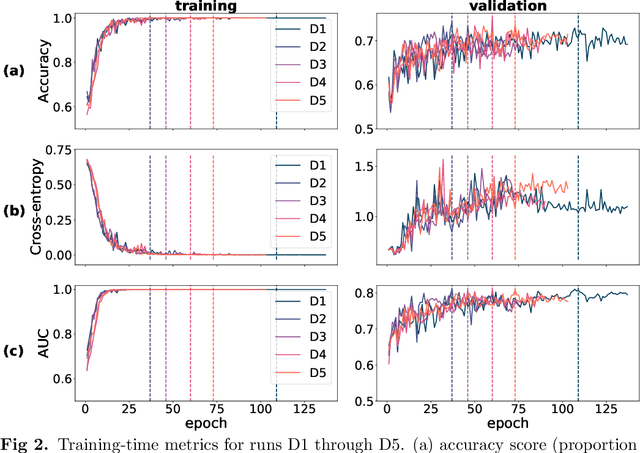
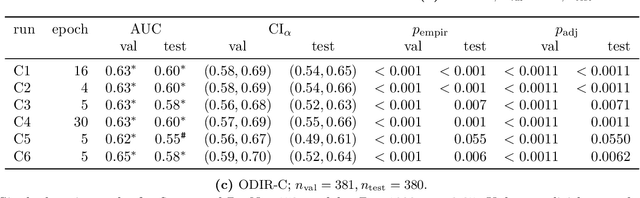
Abstract:Deep learning has seen tremendous interest in medical imaging, particularly in the use of convolutional neural networks (CNNs) for developing automated diagnostic tools. The facility of its non-invasive acquisition makes retinal fundus imaging amenable to such automated approaches. Recent work in analyzing fundus images using CNNs relies on access to massive data for training and validation - hundreds of thousands of images. However, data residency and data privacy restrictions stymie the applicability of this approach in medical settings where patient confidentiality is a mandate. Here, we showcase results for the performance of DL on small datasets to classify patient sex from fundus images - a trait thought not to be present or quantifiable in fundus images until recently. We fine-tune a Resnet-152 model whose last layer has been modified for binary classification. In several experiments, we assess performance in the small dataset context using one private (DOVS) and one public (ODIR) data source. Our models, developed using approximately 2500 fundus images, achieved test AUC scores of up to 0.72 (95% CI: [0.67, 0.77]). This corresponds to a mere 25% decrease in performance despite a nearly 1000-fold decrease in the dataset size compared to prior work in the literature. Even with a hard task like sex categorization from retinal images, we find that classification is possible with very small datasets. Additionally, we perform domain adaptation experiments between DOVS and ODIR; explore the effect of data curation on training and generalizability; and investigate model ensembling to maximize CNN classifier performance in the context of small development datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge