Iñaki Soto-Rey
DeepGleason: a System for Automated Gleason Grading of Prostate Cancer using Deep Neural Networks
Mar 25, 2024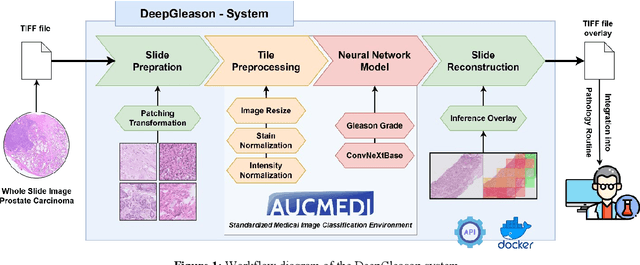
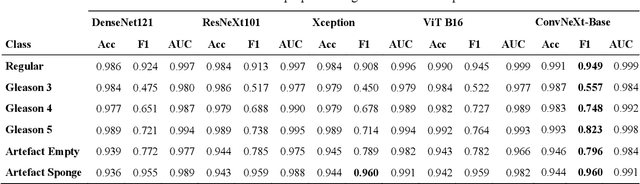
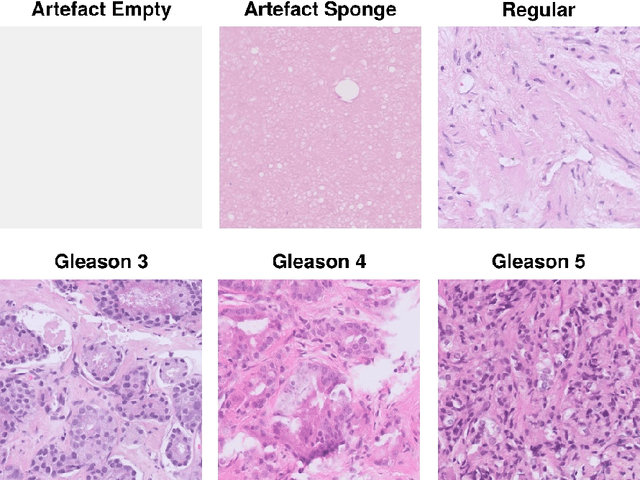
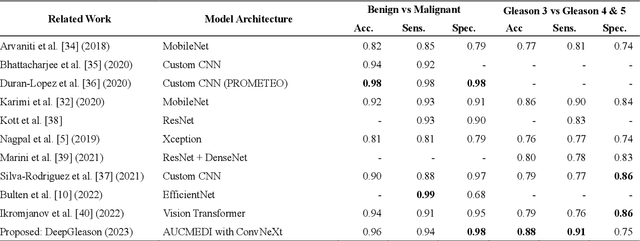
Abstract:Advances in digital pathology and artificial intelligence (AI) offer promising opportunities for clinical decision support and enhancing diagnostic workflows. Previous studies already demonstrated AI's potential for automated Gleason grading, but lack state-of-the-art methodology and model reusability. To address this issue, we propose DeepGleason: an open-source deep neural network based image classification system for automated Gleason grading using whole-slide histopathology images from prostate tissue sections. Implemented with the standardized AUCMEDI framework, our tool employs a tile-wise classification approach utilizing fine-tuned image preprocessing techniques in combination with a ConvNeXt architecture which was compared to various state-of-the-art architectures. The neural network model was trained and validated on an in-house dataset of 34,264 annotated tiles from 369 prostate carcinoma slides. We demonstrated that DeepGleason is capable of highly accurate and reliable Gleason grading with a macro-averaged F1-score of 0.806, AUC of 0.991, and Accuracy of 0.974. The internal architecture comparison revealed that the ConvNeXt model was superior performance-wise on our dataset to established and other modern architectures like transformers. Furthermore, we were able to outperform the current state-of-the-art in tile-wise fine-classification with a sensitivity and specificity of 0.94 and 0.98 for benign vs malignant detection as well as of 0.91 and 0.75 for Gleason 3 vs Gleason 4 & 5 classification, respectively. Our tool contributes to the wider adoption of AI-based Gleason grading within the research community and paves the way for broader clinical application of deep learning models in digital pathology. DeepGleason is open-source and publicly available for research application in the following Git repository: https://github.com/frankkramer-lab/DeepGleason.
Assessing the Performance of Deep Learning for Automated Gleason Grading in Prostate Cancer
Mar 25, 2024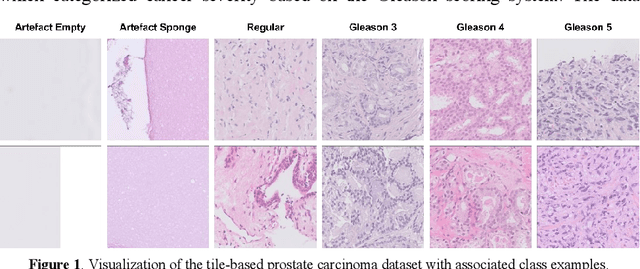
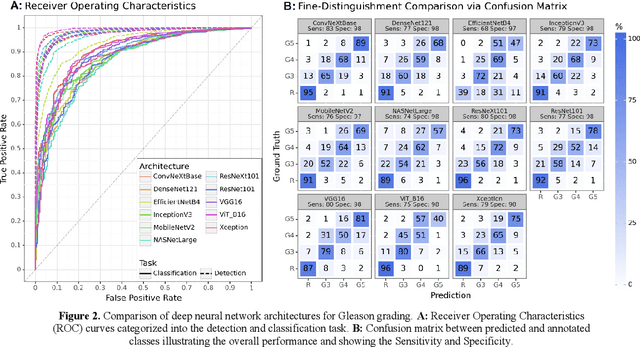
Abstract:Prostate cancer is a dominant health concern calling for advanced diagnostic tools. Utilizing digital pathology and artificial intelligence, this study explores the potential of 11 deep neural network architectures for automated Gleason grading in prostate carcinoma focusing on comparing traditional and recent architectures. A standardized image classification pipeline, based on the AUCMEDI framework, facilitated robust evaluation using an in-house dataset consisting of 34,264 annotated tissue tiles. The results indicated varying sensitivity across architectures, with ConvNeXt demonstrating the strongest performance. Notably, newer architectures achieved superior performance, even though with challenges in differentiating closely related Gleason grades. The ConvNeXt model was capable of learning a balance between complexity and generalizability. Overall, this study lays the groundwork for enhanced Gleason grading systems, potentially improving diagnostic efficiency for prostate cancer.
MISm: A Medical Image Segmentation Metric for Evaluation of weak labeled Data
Oct 24, 2022Abstract:Performance measures are an important tool for assessing and comparing different medical image segmentation algorithms. Unfortunately, the current measures have their weaknesses when it comes to assessing certain edge cases. These limitations arouse when images with a very small region of interest or without a region of interest at all are assessed. As a solution for these limitations, we propose a new medical image segmentation metric: MISm. To evaluate MISm, the popular metrics in the medical image segmentation and MISm were compared using images of magnet resonance tomography from several scenarios. In order to allow application in the community and reproducibility of experimental results, we included MISm in the publicly available evaluation framework MISeval: https://github.com/frankkramer-lab/miseval/tree/master/miseval
Towards a Guideline for Evaluation Metrics in Medical Image Segmentation
Feb 10, 2022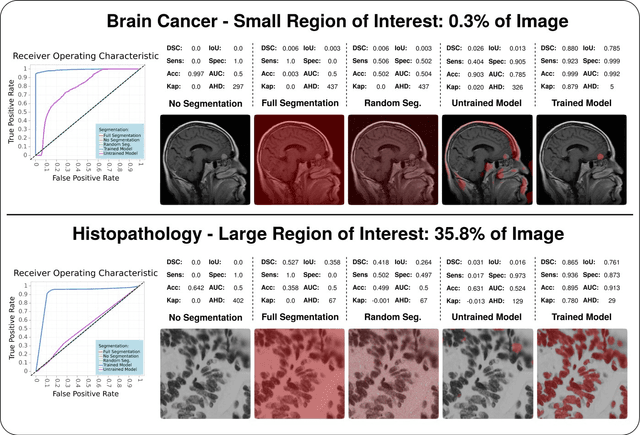
Abstract:In the last decade, research on artificial intelligence has seen rapid growth with deep learning models, especially in the field of medical image segmentation. Various studies demonstrated that these models have powerful prediction capabilities and achieved similar results as clinicians. However, recent studies revealed that the evaluation in image segmentation studies lacks reliable model performance assessment and showed statistical bias by incorrect metric implementation or usage. Thus, this work provides an overview and interpretation guide on the following metrics for medical image segmentation evaluation in binary as well as multi-class problems: Dice similarity coefficient, Jaccard, Sensitivity, Specificity, Rand index, ROC curves, Cohen's Kappa, and Hausdorff distance. As a summary, we propose a guideline for standardized medical image segmentation evaluation to improve evaluation quality, reproducibility, and comparability in the research field.
An Analysis on Ensemble Learning optimized Medical Image Classification with Deep Convolutional Neural Networks
Jan 27, 2022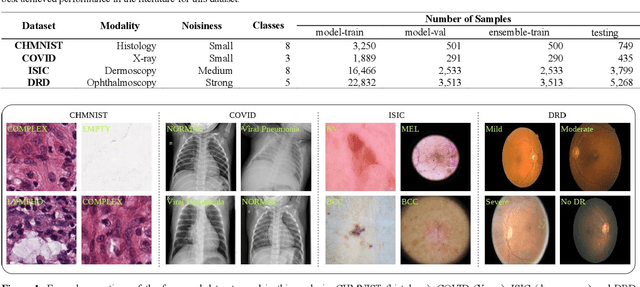
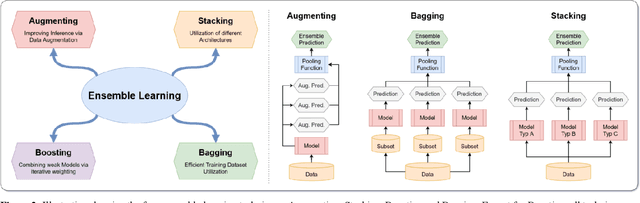
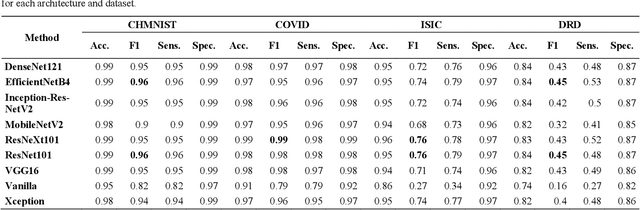
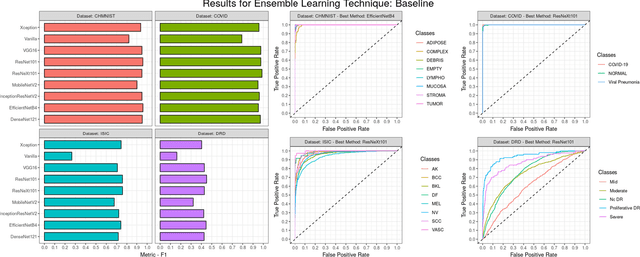
Abstract:Novel and high-performance medical image classification pipelines are heavily utilizing ensemble learning strategies. The idea of ensemble learning is to assemble diverse models or multiple predictions and, thus, boost prediction performance. However, it is still an open question to what extent as well as which ensemble learning strategies are beneficial in deep learning based medical image classification pipelines. In this work, we proposed a reproducible medical image classification pipeline for analyzing the performance impact of the following ensemble learning techniques: Augmenting, Stacking, and Bagging. The pipeline consists of state-of-the-art preprocessing and image augmentation methods as well as 9 deep convolution neural network architectures. It was applied on four popular medical imaging datasets with varying complexity. Furthermore, 12 pooling functions for combining multiple predictions were analyzed, ranging from simple statistical functions like unweighted averaging up to more complex learning-based functions like support vector machines. Our results revealed that Stacking achieved the largest performance gain of up to 13% F1-score increase. Augmenting showed consistent improvement capabilities by up to 4% and is also applicable to single model based pipelines. Cross-validation based Bagging demonstrated to be the most complex ensemble learning method, which resulted in an F1-score decrease in all analyzed datasets (up to -10%). Furthermore, we demonstrated that simple statistical pooling functions are equal or often even better than more complex pooling functions. We concluded that the integration of Stacking and Augmentation ensemble learning techniques is a powerful method for any medical image classification pipeline to improve robustness and boost performance.
MISeval: a Metric Library for Medical Image Segmentation Evaluation
Jan 23, 2022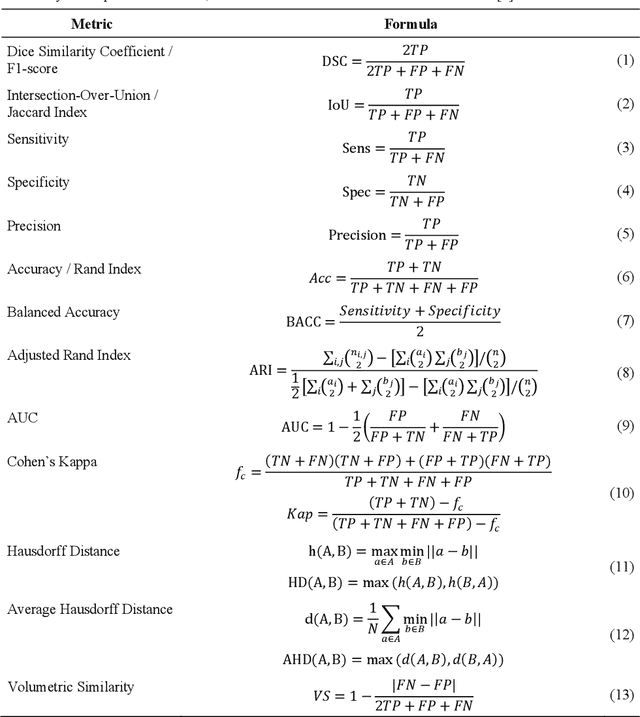
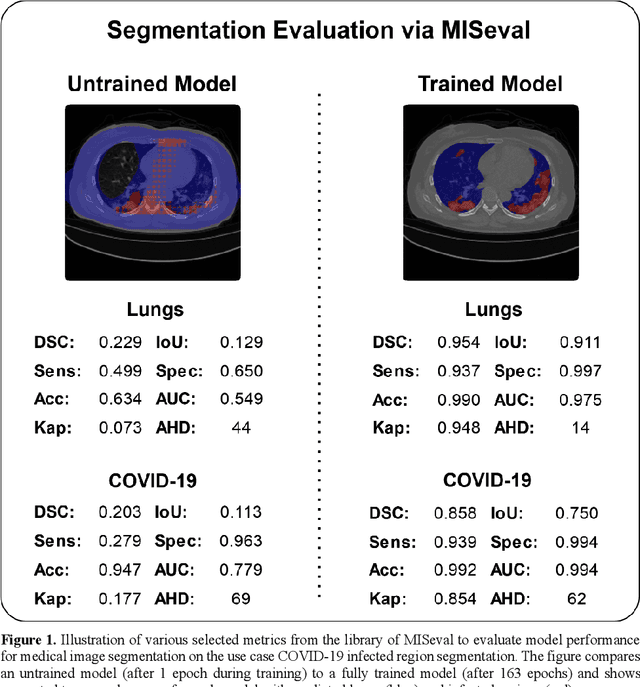
Abstract:Correct performance assessment is crucial for evaluating modern artificial intelligence algorithms in medicine like deep-learning based medical image segmentation models. However, there is no universal metric library in Python for standardized and reproducible evaluation. Thus, we propose our open-source publicly available Python package MISeval: a metric library for Medical Image Segmentation Evaluation. The implemented metrics can be intuitively used and easily integrated into any performance assessment pipeline. The package utilizes modern CI/CD strategies to ensure functionality and stability. MISeval is available from PyPI (miseval) and GitHub: https://github.com/frankkramer-lab/miseval.
Assessing the Role of Random Forests in Medical Image Segmentation
Mar 30, 2021



Abstract:Neural networks represent a field of research that can quickly achieve very good results in the field of medical image segmentation using a GPU. A possible way to achieve good results without GPUs are random forests. For this purpose, two random forest approaches were compared with a state-of-the-art deep convolutional neural network. To make the comparison the PhC-C2DH-U373 and the retinal imaging datasets were used. The evaluation showed that the deep convolutional neutral network achieved the best results. However, one of the random forest approaches also achieved a similar high performance. Our results indicate that random forest approaches are a good alternative to deep convolutional neural networks and, thus, allow the usage of medical image segmentation without a GPU.
Multi-Disease Detection in Retinal Imaging based on Ensembling Heterogeneous Deep Learning Models
Mar 26, 2021


Abstract:Preventable or undiagnosed visual impairment and blindness affect billion of people worldwide. Automated multi-disease detection models offer great potential to address this problem via clinical decision support in diagnosis. In this work, we proposed an innovative multi-disease detection pipeline for retinal imaging which utilizes ensemble learning to combine the predictive capabilities of several heterogeneous deep convolutional neural network models. Our pipeline includes state-of-the-art strategies like transfer learning, class weighting, real-time image augmentation and Focal loss utilization. Furthermore, we integrated ensemble learning techniques like heterogeneous deep learning models, bagging via 5-fold cross-validation and stacked logistic regression models. Through internal and external evaluation, we were able to validate and demonstrate high accuracy and reliability of our pipeline, as well as the comparability with other state-of-the-art pipelines for retinal disease prediction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge