Hoshik Lee
Image-to-Graph Transformers for Chemical Structure Recognition
Feb 19, 2022
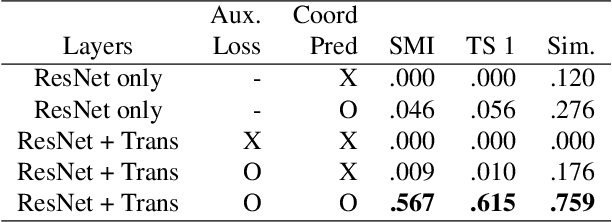
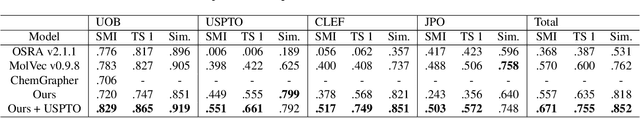
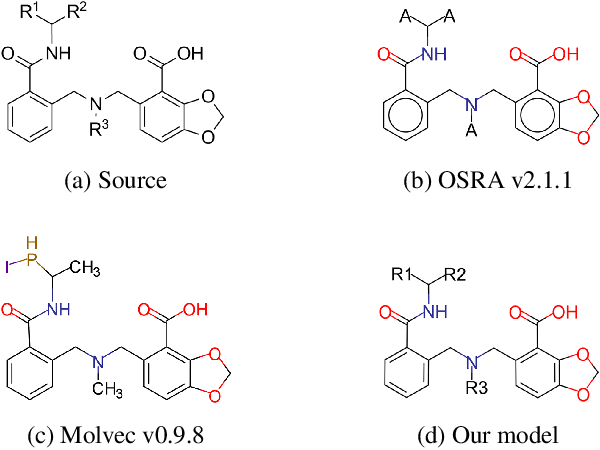
Abstract:For several decades, chemical knowledge has been published in written text, and there have been many attempts to make it accessible, for example, by transforming such natural language text to a structured format. Although the discovered chemical itself commonly represented in an image is the most important part, the correct recognition of the molecular structure from the image in literature still remains a hard problem since they are often abbreviated to reduce the complexity and drawn in many different styles. In this paper, we present a deep learning model to extract molecular structures from images. The proposed model is designed to transform the molecular image directly into the corresponding graph, which makes it capable of handling non-atomic symbols for abbreviations. Also, by end-to-end learning approach it can fully utilize many open image-molecule pair data from various sources, and hence it is more robust to image style variation than other tools. The experimental results show that the proposed model outperforms the existing models with 17.1 % and 12.8 % relative improvement for well-known benchmark datasets and large molecular images that we collected from literature, respectively.
Graph-Aware Transformer: Is Attention All Graphs Need?
Jun 09, 2020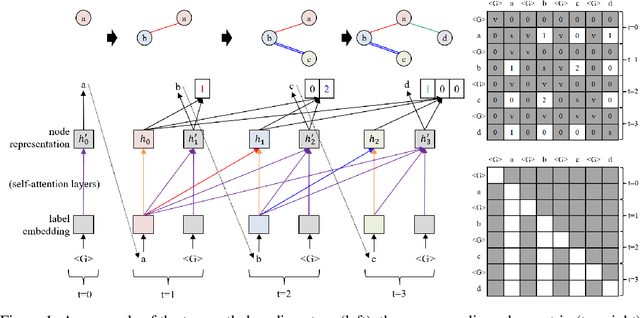

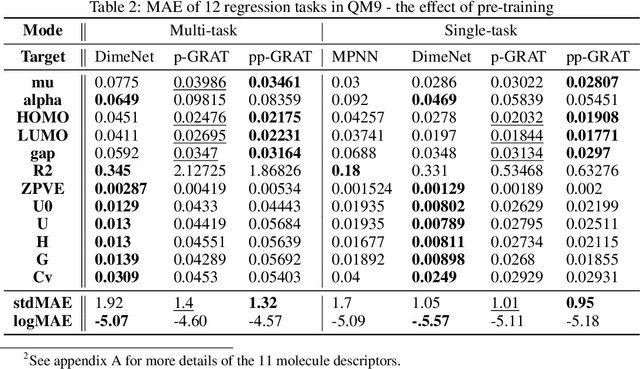
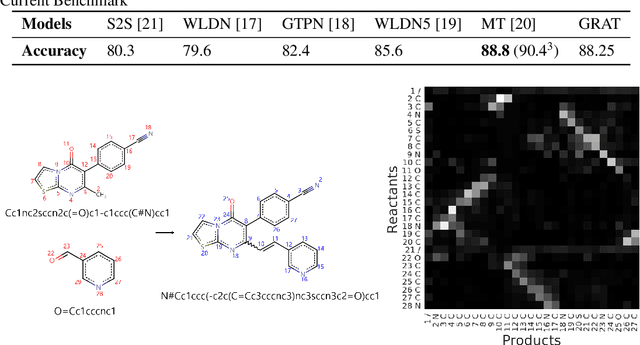
Abstract:Graphs are the natural data structure to represent relational and structural information in many domains. To cover the broad range of graph-data applications including graph classification as well as graph generation, it is desirable to have a general and flexible model consisting of an encoder and a decoder that can handle graph data. Although the representative encoder-decoder model, Transformer, shows superior performance in various tasks especially of natural language processing, it is not immediately available for graphs due to their non-sequential characteristics. To tackle this incompatibility, we propose GRaph-Aware Transformer (GRAT), the first Transformer-based model which can encode and decode whole graphs in end-to-end fashion. GRAT is featured with a self-attention mechanism adaptive to the edge information and an auto-regressive decoding mechanism based on the two-path approach consisting of sub-graph encoding path and node-and-edge generation path for each decoding step. We empirically evaluated GRAT on multiple setups including encoder-based tasks such as molecule property predictions on QM9 datasets and encoder-decoder-based tasks such as molecule graph generation in the organic molecule synthesis domain. GRAT has shown very promising results including state-of-the-art performance on 4 regression tasks in QM9 benchmark.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge