Heather D. Couture
Deep Learning-Based Prediction of Molecular Tumor Biomarkers from H&E: A Practical Review
Nov 27, 2022Abstract:Molecular and genomic properties are critical in selecting cancer treatments to target individual tumors, particularly for immunotherapy. However, the methods to assess such properties are expensive, time-consuming, and often not routinely performed. Applying machine learning to H&E images can provide a more cost-effective screening method. Dozens of studies over the last few years have demonstrated that a variety of molecular biomarkers can be predicted from H&E alone using the advancements of deep learning: molecular alterations, genomic subtypes, protein biomarkers, and even the presence of viruses. This article reviews the diverse applications across cancer types and the methodology to train and validate these models on whole slide images. From bottom-up to pathologist-driven to hybrid approaches, the leading trends include a variety of weakly supervised deep learning-based approaches, as well as mechanisms for training strongly supervised models in select situations. While results of these algorithms look promising, some challenges still persist, including small training sets, rigorous validation, and model explainability. Biomarker prediction models may yield a screening method to determine when to run molecular tests or an alternative when molecular tests are not possible. They also create new opportunities in quantifying intratumoral heterogeneity and predicting patient outcomes.
Deep Multi-View Learning via Task-Optimal CCA
Jul 17, 2019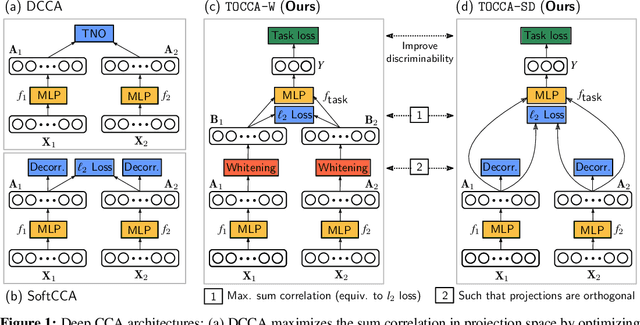
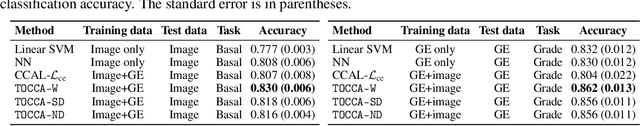
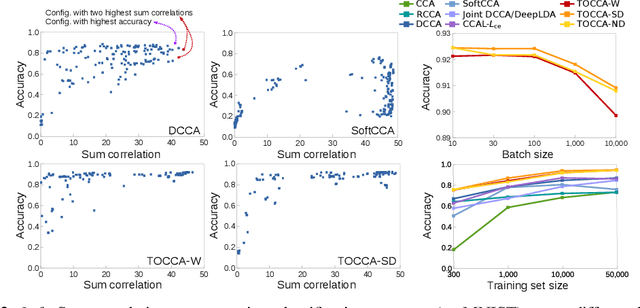
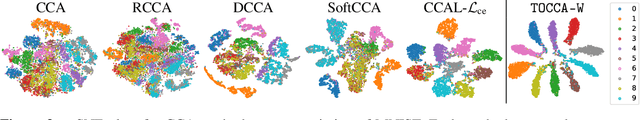
Abstract:Canonical Correlation Analysis (CCA) is widely used for multimodal data analysis and, more recently, for discriminative tasks such as multi-view learning; however, it makes no use of class labels. Recent CCA methods have started to address this weakness but are limited in that they do not simultaneously optimize the CCA projection for discrimination and the CCA projection itself, or they are linear only. We address these deficiencies by simultaneously optimizing a CCA-based and a task objective in an end-to-end manner. Together, these two objectives learn a non-linear CCA projection to a shared latent space that is highly correlated and discriminative. Our method shows a significant improvement over previous state-of-the-art (including deep supervised approaches) for cross-view classification, regularization with a second view, and semi-supervised learning on real data.
Multiple Instance Learning for Heterogeneous Images: Training a CNN for Histopathology
Jun 13, 2018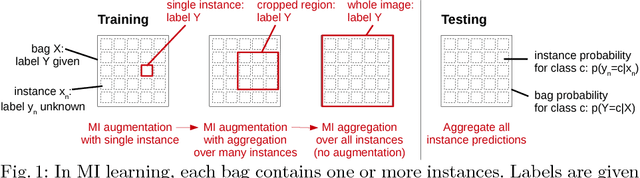
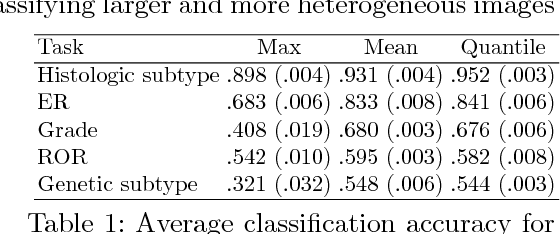
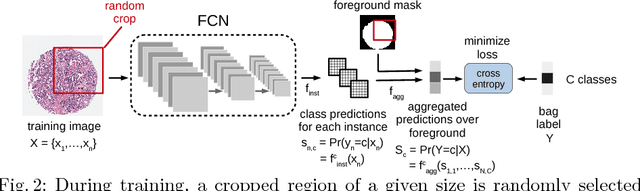
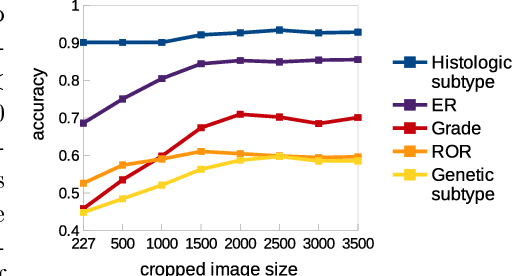
Abstract:Multiple instance (MI) learning with a convolutional neural network enables end-to-end training in the presence of weak image-level labels. We propose a new method for aggregating predictions from smaller regions of the image into an image-level classification by using the quantile function. The quantile function provides a more complete description of the heterogeneity within each image, improving image-level classification. We also adapt image augmentation to the MI framework by randomly selecting cropped regions on which to apply MI aggregation during each epoch of training. This provides a mechanism to study the importance of MI learning. We validate our method on five different classification tasks for breast tumor histology and provide a visualization method for interpreting local image classifications that could lead to future insights into tumor heterogeneity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge