Guy Hagen
Raman spectroscopy in open world learning settings using the Objectosphere approach
Nov 11, 2021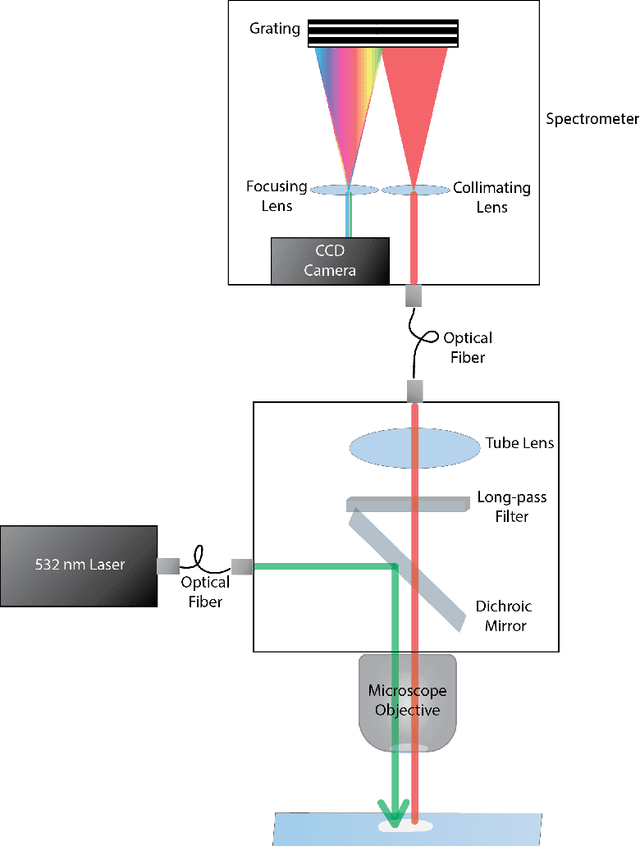
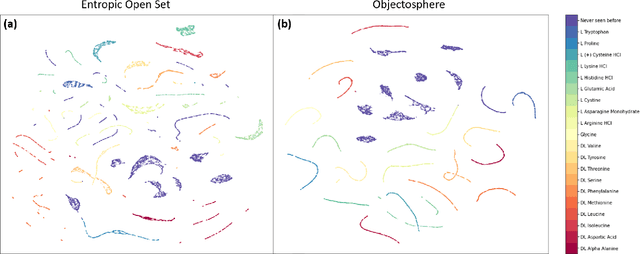
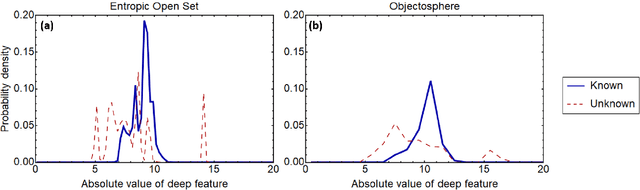
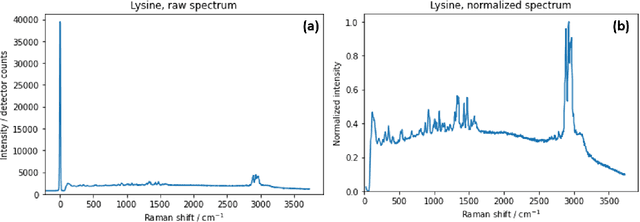
Abstract:Raman spectroscopy in combination with machine learning has significant promise for applications in clinical settings as a rapid, sensitive, and label-free identification method. These approaches perform well in classifying data that contains classes that occur during the training phase. However, in practice, there are always substances whose spectra have not yet been taken or are not yet known and when the input data are far from the training set and include new classes that were not seen at the training stage, a significant number of false positives are recorded which limits the clinical relevance of these algorithms. Here we show that these obstacles can be overcome by implementing recently introduced Entropic Open Set and Objectosphere loss functions. To demonstrate the efficiency of this approach, we compiled a database of Raman spectra of 40 chemical classes separating them into 20 biologically relevant classes comprised of amino acids, 10 irrelevant classes comprised of bio-related chemicals, and 10 classes that the Neural Network has not seen before, comprised of a variety of other chemicals. We show that this approach enables the network to effectively identify the unknown classes while preserving high accuracy on the known ones, dramatically reducing the number of false positives while preserving high accuracy on the known classes, which will allow this technique to bridge the gap between laboratory experiments and clinical applications.
Self-Supervised Poisson-Gaussian Denoising
Feb 21, 2020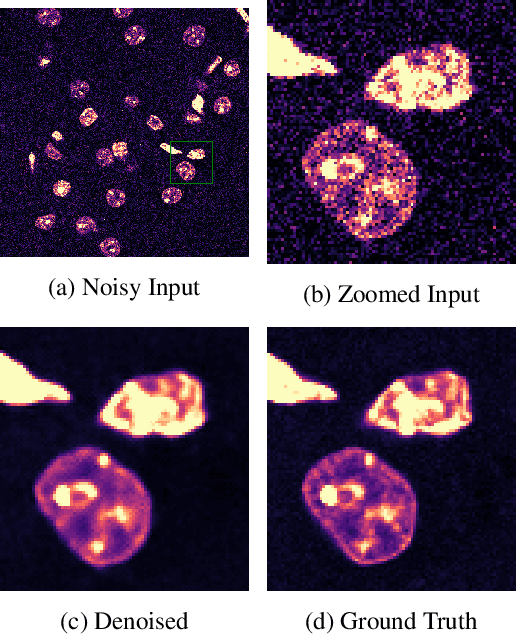
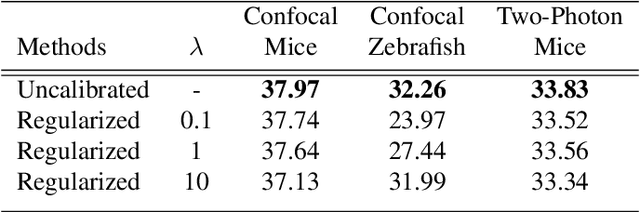
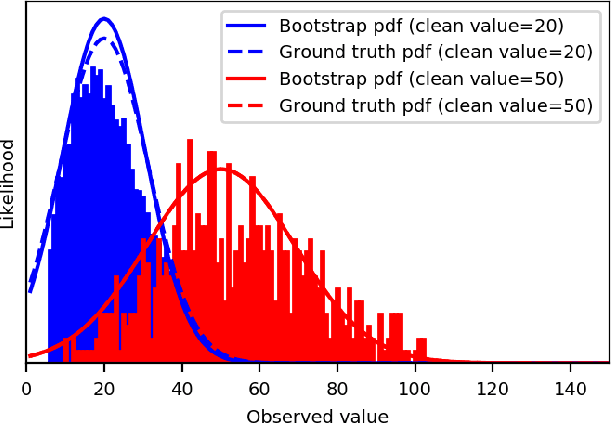
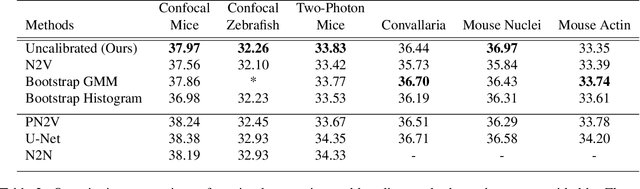
Abstract:We extend the blindspot model for self-supervised denoising to handle Poisson-Gaussian noise and introduce an improved training scheme that avoids hyperparameters and adapts the denoiser to the test data. Self-supervised models for denoising learn to denoise from only noisy data and do not require corresponding clean images, which are difficult or impossible to acquire in some application areas of interest such as low-light microscopy. We introduce a new training strategy to handle Poisson-Gaussian noise which is the standard noise model for microscope images. Our new strategy eliminates hyperparameters from the loss function, which is important in a self-supervised regime where no ground truth data is available to guide hyperparameter tuning. We show how our denoiser can be adapted to the test data to improve performance. Our evaluation on a microscope image denoising benchmark validates our approach.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge