Kelly McNear
Enhancing Open-World Bacterial Raman Spectra Identification by Feature Regularization for Improved Resilience against Unknown Classes
Oct 19, 2023Abstract:The combination of Deep Learning techniques and Raman spectroscopy shows great potential offering precise and prompt identification of pathogenic bacteria in clinical settings. However, the traditional closed-set classification approaches assume that all test samples belong to one of the known pathogens, and their applicability is limited since the clinical environment is inherently unpredictable and dynamic, unknown or emerging pathogens may not be included in the available catalogs. We demonstrate that the current state-of-the-art Neural Networks identifying pathogens through Raman spectra are vulnerable to unknown inputs, resulting in an uncontrollable false positive rate. To address this issue, first, we developed a novel ensemble of ResNet architectures combined with the attention mechanism which outperforms existing closed-world methods, achieving an accuracy of $87.8 \pm 0.1\%$ compared to the best available model's accuracy of $86.7 \pm 0.4\%$. Second, through the integration of feature regularization by the Objectosphere loss function, our model achieves both high accuracy in identifying known pathogens from the catalog and effectively separates unknown samples drastically reducing the false positive rate. Finally, the proposed feature regularization method during training significantly enhances the performance of out-of-distribution detectors during the inference phase improving the reliability of the detection of unknown classes. Our novel algorithm for Raman spectroscopy enables the detection of unknown, uncatalogued, and emerging pathogens providing the flexibility to adapt to future pathogens that may emerge, and has the potential to improve the reliability of Raman-based solutions in dynamic operating environments where accuracy is critical, such as public safety applications.
Raman spectroscopy in open world learning settings using the Objectosphere approach
Nov 11, 2021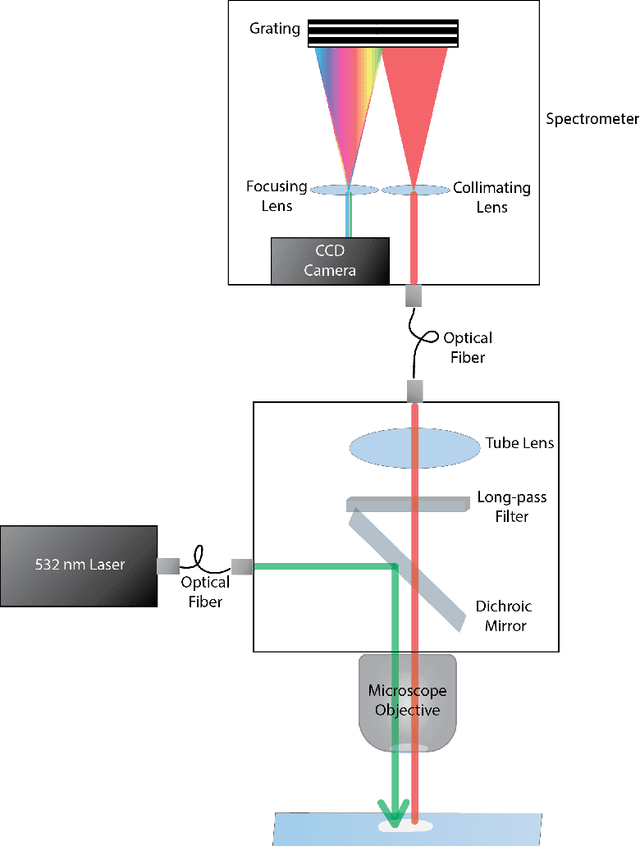
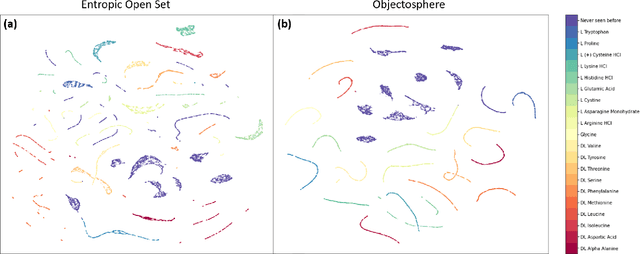
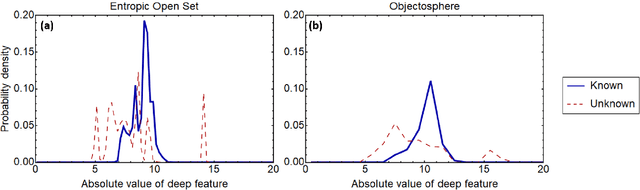
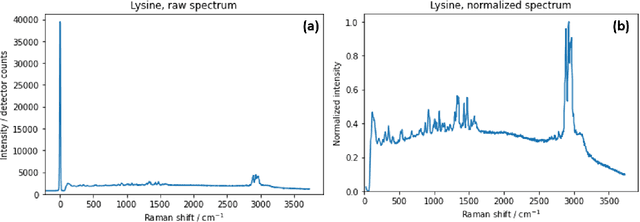
Abstract:Raman spectroscopy in combination with machine learning has significant promise for applications in clinical settings as a rapid, sensitive, and label-free identification method. These approaches perform well in classifying data that contains classes that occur during the training phase. However, in practice, there are always substances whose spectra have not yet been taken or are not yet known and when the input data are far from the training set and include new classes that were not seen at the training stage, a significant number of false positives are recorded which limits the clinical relevance of these algorithms. Here we show that these obstacles can be overcome by implementing recently introduced Entropic Open Set and Objectosphere loss functions. To demonstrate the efficiency of this approach, we compiled a database of Raman spectra of 40 chemical classes separating them into 20 biologically relevant classes comprised of amino acids, 10 irrelevant classes comprised of bio-related chemicals, and 10 classes that the Neural Network has not seen before, comprised of a variety of other chemicals. We show that this approach enables the network to effectively identify the unknown classes while preserving high accuracy on the known ones, dramatically reducing the number of false positives while preserving high accuracy on the known classes, which will allow this technique to bridge the gap between laboratory experiments and clinical applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge