Gregory Henselman-Petrusek
Computable Stability for Persistence Rank Function Machine Learning
Jul 06, 2023Abstract:Persistent homology barcodes and diagrams are a cornerstone of topological data analysis. Widely used in many real data settings, they relate variation in topological information (as measured by cellular homology) with variation in data, however, they are challenging to use in statistical settings due to their complex geometric structure. In this paper, we revisit the persistent homology rank function -- an invariant measure of ``shape" that was introduced before barcodes and persistence diagrams and captures the same information in a form that is more amenable to data and computation. In particular, since they are functions, techniques from functional data analysis -- a domain of statistics adapted for functions -- apply directly to persistent homology when represented by rank functions. Rank functions, however, have been less popular than barcodes because they face the challenge that stability -- a property that is crucial to validate their use in data analysis -- is difficult to guarantee, mainly due to metric concerns on rank function space. However, rank functions extend more naturally to the increasingly popular and important case of multiparameter persistent homology. In this paper, we study the performance of rank functions in functional inferential statistics and machine learning on both simulated and real data, and in both single and multiparameter persistent homology. We find that the use of persistent homology captured by rank functions offers a clear improvement over existing approaches. We then provide theoretical justification for our numerical experiments and applications to data by deriving several stability results for single- and multiparameter persistence rank functions under various metrics with the underlying aim of computational feasibility and interpretability.
Minimal Cycle Representatives in Persistent Homology using Linear Programming: an Empirical Study with User's Guide
May 18, 2021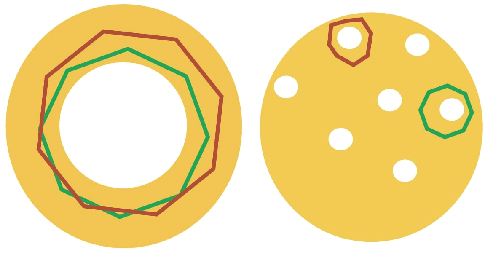
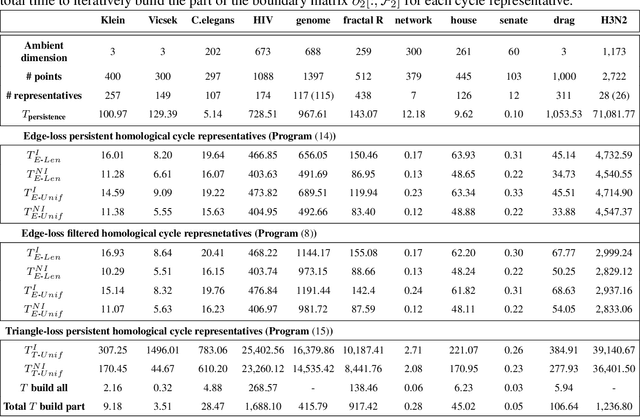
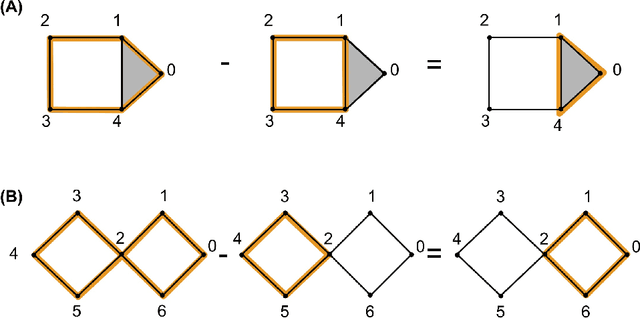
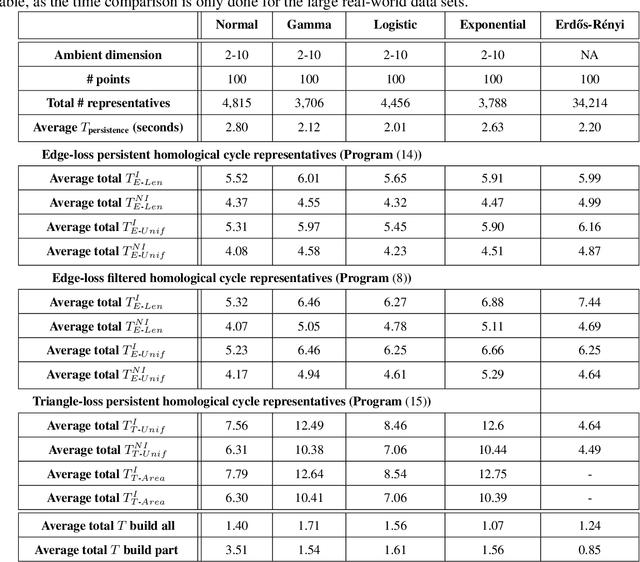
Abstract:Cycle representatives of persistent homology classes can be used to provide descriptions of topological features in data. However, the non-uniqueness of these representatives creates ambiguity and can lead to many different interpretations of the same set of classes. One approach to solving this problem is to optimize the choice of representative against some measure that is meaningful in the context of the data. In this work, we provide a study of the effectiveness and computational cost of several $\ell_1$-minimization optimization procedures for constructing homological cycle bases for persistent homology with rational coefficients in dimension one, including uniform-weighted and length-weighted edge-loss algorithms as well as uniform-weighted and area-weighted triangle-loss algorithms. We conduct these optimizations via standard linear programming methods, applying general-purpose solvers to optimize over column bases of simplicial boundary matrices. Our key findings are: (i) optimization is effective in reducing the size of cycle representatives, (ii) the computational cost of optimizing a basis of cycle representatives exceeds the cost of computing such a basis in most data sets we consider, (iii) the choice of linear solvers matters a lot to the computation time of optimizing cycles, (iv) the computation time of solving an integer program is not significantly longer than the computation time of solving a linear program for most of the cycle representatives, using the Gurobi linear solver, (v) strikingly, whether requiring integer solutions or not, we almost always obtain a solution with the same cost and almost all solutions found have entries in {-1, 0, 1} and therefore, are also solutions to a restricted $\ell_0$ optimization problem, and (vi) we obtain qualitatively different results for generators in Erd\H{o}s-R\'enyi random clique complexes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge