Gracia Sánchez
Permutation-based multi-objective evolutionary feature selection for high-dimensional data
Jan 24, 2025Abstract:Feature selection is a critical step in the analysis of high-dimensional data, where the number of features often vastly exceeds the number of samples. Effective feature selection not only improves model performance and interpretability but also reduces computational costs and mitigates the risk of overfitting. In this context, we propose a novel feature selection method for high-dimensional data, based on the well-known permutation feature importance approach, but extending it to evaluate subsets of attributes rather than individual features. This extension more effectively captures how interactions among features influence model performance. The proposed method employs a multi-objective evolutionary algorithm to search for candidate feature subsets, with the objectives of maximizing the degradation in model performance when the selected features are shuffled, and minimizing the cardinality of the feature subset. The effectiveness of our method has been validated on a set of 24 publicly available high-dimensional datasets for classification and regression tasks, and compared against 9 well-established feature selection methods designed for high-dimensional problems, including the conventional permutation feature importance method. The results demonstrate the ability of our approach in balancing accuracy and computational efficiency, providing a powerful tool for feature selection in complex, high-dimensional datasets.
Multivariate feature ranking of gene expression data
Nov 16, 2021
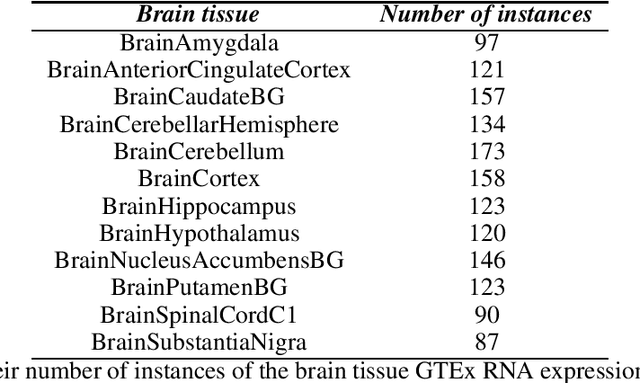


Abstract:Gene expression datasets are usually of high dimensionality and therefore require efficient and effective methods for identifying the relative importance of their attributes. Due to the huge size of the search space of the possible solutions, the attribute subset evaluation feature selection methods tend to be not applicable, so in these scenarios feature ranking methods are used. Most of the feature ranking methods described in the literature are univariate methods, so they do not detect interactions between factors. In this paper we propose two new multivariate feature ranking methods based on pairwise correlation and pairwise consistency, which we have applied in three gene expression classification problems. We statistically prove that the proposed methods outperform the state of the art feature ranking methods Clustering Variation, Chi Squared, Correlation, Information Gain, ReliefF and Significance, as well as feature selection methods of attribute subset evaluation based on correlation and consistency with multi-objective evolutionary search strategy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge