Multivariate feature ranking of gene expression data
Paper and Code
Nov 16, 2021
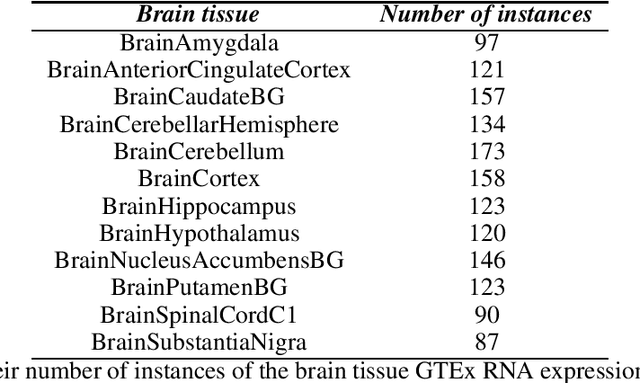


Gene expression datasets are usually of high dimensionality and therefore require efficient and effective methods for identifying the relative importance of their attributes. Due to the huge size of the search space of the possible solutions, the attribute subset evaluation feature selection methods tend to be not applicable, so in these scenarios feature ranking methods are used. Most of the feature ranking methods described in the literature are univariate methods, so they do not detect interactions between factors. In this paper we propose two new multivariate feature ranking methods based on pairwise correlation and pairwise consistency, which we have applied in three gene expression classification problems. We statistically prove that the proposed methods outperform the state of the art feature ranking methods Clustering Variation, Chi Squared, Correlation, Information Gain, ReliefF and Significance, as well as feature selection methods of attribute subset evaluation based on correlation and consistency with multi-objective evolutionary search strategy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge